Alterations in the epithelial stem cell compartment could contribute to permanent changes in the mucosa of patients with ulcerative colitis
- PMID: 27803115
- PMCID: PMC5749340
- DOI: 10.1136/gutjnl-2016-312609
Alterations in the epithelial stem cell compartment could contribute to permanent changes in the mucosa of patients with ulcerative colitis
Abstract
Objective: UC is a chronic inflammatory disease of the colonic mucosa. Growing evidence supports a role for epithelial cell defects in driving pathology. Moreover, long-lasting changes in the epithelial barrier have been reported in quiescent UC. Our aim was to investigate whether epithelial cell defects could originate from changes in the epithelial compartment imprinted by the disease.
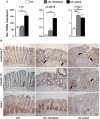
Design: Epithelial organoid cultures (EpOCs) were expanded ex vivo from the intestinal crypts of non-IBD controls and patients with UC. EpOCs were induced to differentiate (d-EpOCs), and the total RNA was extracted for microarray and quantitative real-time PCR (qPCR) analyses. Whole intestinal samples were used to determine mRNA expression by qPCR, or protein localisation by immunostaining.
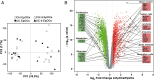
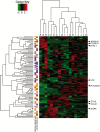
Results: EpOCs from patients with UC maintained self-renewal potential and the capability to give rise to differentiated epithelial cell lineages comparable with control EpOCs. Nonetheless, a group of genes was differentially regulated in the EpOCs and d-EpOCs of patients with UC, including genes associated with antimicrobial defence (ie, LYZ, PLA2G2A), with secretory (ie, ZG16, CLCA1) and absorptive (ie, AQP8, MUC12) functions, and with a gastric phenotype (ie, ANXA10, CLDN18 and LYZ). A high rate of concordance was found in the expression profiles of the organoid cultures and whole colonic tissues from patients with UC.
Conclusions: Permanent changes in the colonic epithelium of patients with UC could be promoted by alterations imprinted in the stem cell compartment. These changes may contribute to perpetuation of the disease.
Keywords: GENE EXPRESSION; INTESTINAL EPITHELIUM; STEM CELLS; ULCERATIVE COLITIS.
Published by the BMJ Publishing Group Limited. For permission to use (where not already granted under a licence) please go to http://www.bmj.com/company/products-services/rights-and-licensing/.
Conflict of interest statement
Competing interests: None declared.
Figures




Comment in
-
Epithelial organoid cultures from patients with ulcerative colitis and Crohn's disease: a truly long-term model to study the molecular basis for inflammatory bowel disease?Gut. 2017 Dec;66(12):2193-2195. doi: 10.1136/gutjnl-2016-313667. Epub 2017 Feb 3. Gut. 2017. PMID: 28159838 No abstract available.
-
Intestinal organoid culture model is a valuable system to study epithelial barrier function in IBD.Gut. 2018 Oct;67(10):1905-1906. doi: 10.1136/gutjnl-2017-315685. Epub 2017 Dec 5. Gut. 2018. PMID: 29208677 No abstract available.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
