S2 from equine infectious anemia virus is an infectivity factor which counteracts the retroviral inhibitors SERINC5 and SERINC3
- PMID: 27803322
- PMCID: PMC5135340
- DOI: 10.1073/pnas.1612044113
S2 from equine infectious anemia virus is an infectivity factor which counteracts the retroviral inhibitors SERINC5 and SERINC3
Abstract
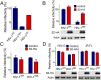
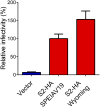
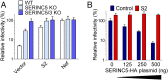
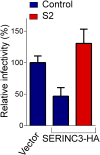
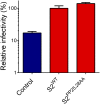
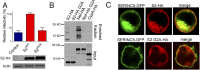
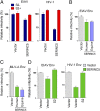
The lentivirus equine infectious anemia virus (EIAV) encodes the small protein S2, a pathogenic determinant that is important for virus replication and disease progression in horses. No molecular function had been linked to this accessory protein. We report that S2 can replace the activity of Negative factor (Nef) in HIV-1 infectivity, being required to antagonize the inhibitory activity of Serine incorporator (SERINC) proteins on Nef-defective HIV-1. Like Nef, S2 excludes SERINC5 from virus particles and requires an ExxxLL motif predicted to recruit the clathrin adaptor, Adaptor protein 2 (AP2). Accordingly, functional endocytic machinery is essential for S2-mediated infectivity enhancement, and S2-mediated enhancement is impaired by inhibitors of clathrin-mediated endocytosis. In addition to retargeting SERINC5 to a late endosomal compartment, S2 promotes host factor degradation. Emphasizing the similarity with Nef, we show that S2 is myristoylated, and, as is compatible with a crucial role in posttranslational modification, its N-terminal glycine is required for anti-SERINC5 activity. EIAV-derived vectors devoid of S2 are less susceptible than HIV-1 to the inhibitory effect of both human and equine SERINC5. We then identified the envelope glycoprotein of EIAV as a determinant that also modulates retroviral susceptibility to SERINC5, indicating that EIAV has a bimodal ability to counteract the host factor. S2 shares no sequence homology with other retroviral factors known to counteract SERINC5. Like the primate lentivirus Nef and the gammaretrovirus glycoGag, the accessory protein from EIAV is an example of a retroviral virulence determinant that independently evolved SERINC5-antagonizing activity. SERINC5 therefore plays a critical role in the interaction of the host with diverse retrovirus pathogens.
Keywords: restriction factor; retrovirus; virus infection.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- Kluge SF, Sauter D, Kirchhoff F. SnapShot: Antiviral restriction factors. Cell. 2015;163(3):774–774.e1. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

