Engineering and optimising deaminase fusions for genome editing
- PMID: 27804970
- PMCID: PMC5097136
- DOI: 10.1038/ncomms13330
Engineering and optimising deaminase fusions for genome editing
Erratum in
-
Corrigendum: Engineering and optimising deaminase fusions for genome editing.Nat Commun. 2017 Oct 9;8:16169. doi: 10.1038/ncomms16169. Nat Commun. 2017. PMID: 28991237 Free PMC article.
Abstract
Precise editing is essential for biomedical research and gene therapy. Yet, homology-directed genome modification is limited by the requirements for genomic lesions, homology donors and the endogenous DNA repair machinery. Here we engineered programmable cytidine deaminases and test if we could introduce site-specific cytidine to thymidine transitions in the absence of targeted genomic lesions. Our programmable deaminases effectively convert specific cytidines to thymidines with 13% efficiency in Escherichia coli and 2.5% in human cells. However, off-target deaminations were detected more than 150 bp away from the target site. Moreover, whole genome sequencing revealed that edited bacterial cells did not harbour chromosomal abnormalities but demonstrated elevated global cytidine deamination at deaminase intrinsic binding sites. Therefore programmable deaminases represent a promising genome editing tool in prokaryotes and eukaryotes. Future engineering is required to overcome the processivity and the intrinsic DNA binding affinity of deaminases for safer therapeutic applications.
Conflict of interest statement
L.Y. and G.C. filed the patent related to this manuscript with the application number: US 12/939,505. The remaining authors declare no competing financial interests.
Figures
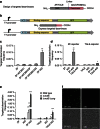


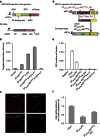
Similar articles
-
Genome-wide target specificities of CRISPR RNA-guided programmable deaminases.Nat Biotechnol. 2017 May;35(5):475-480. doi: 10.1038/nbt.3852. Epub 2017 Apr 10. Nat Biotechnol. 2017. PMID: 28398345
-
A bacterial cytidine deaminase toxin enables CRISPR-free mitochondrial base editing.Nature. 2020 Jul;583(7817):631-637. doi: 10.1038/s41586-020-2477-4. Epub 2020 Jul 8. Nature. 2020. PMID: 32641830 Free PMC article.
-
Increasing the genome-targeting scope and precision of base editing with engineered Cas9-cytidine deaminase fusions.Nat Biotechnol. 2017 Apr;35(4):371-376. doi: 10.1038/nbt.3803. Epub 2017 Feb 13. Nat Biotechnol. 2017. PMID: 28191901 Free PMC article.
-
Generation of Genomic Alteration from Cytidine Deamination.Adv Exp Med Biol. 2018;1044:49-64. doi: 10.1007/978-981-13-0593-1_5. Adv Exp Med Biol. 2018. PMID: 29956291 Review.
-
C-to-U editing and site-directed RNA editing for the correction of genetic mutations.Biosci Trends. 2017 Jul 24;11(3):243-253. doi: 10.5582/bst.2017.01049. Epub 2017 May 8. Biosci Trends. 2017. PMID: 28484188 Review.
Cited by
-
TF-High-Evolutionary: In Vivo Mutagenesis of Gene Regulatory Networks for the Study of the Genetics and Evolution of the Drosophila Regulatory Genome.Mol Biol Evol. 2024 Aug 2;41(8):msae167. doi: 10.1093/molbev/msae167. Mol Biol Evol. 2024. PMID: 39117360 Free PMC article.
-
The Pseudomonas aeruginosa Resistome: Permanent and Transient Antibiotic Resistance, an Overview.Methods Mol Biol. 2024;2721:85-102. doi: 10.1007/978-1-0716-3473-8_7. Methods Mol Biol. 2024. PMID: 37819517
-
Genome-wide target specificities of CRISPR RNA-guided programmable deaminases.Nat Biotechnol. 2017 May;35(5):475-480. doi: 10.1038/nbt.3852. Epub 2017 Apr 10. Nat Biotechnol. 2017. PMID: 28398345
-
The Design and Application of DNA-Editing Enzymes as Base Editors.Annu Rev Biochem. 2023 Jun 20;92:43-79. doi: 10.1146/annurev-biochem-052521-013938. Epub 2023 Apr 5. Annu Rev Biochem. 2023. PMID: 37018843 Free PMC article. Review.
-
Rewritable multi-event analog recording in bacterial and mammalian cells.Science. 2018 Apr 13;360(6385):eaap8992. doi: 10.1126/science.aap8992. Epub 2018 Feb 15. Science. 2018. PMID: 29449507 Free PMC article.
References
-
- Porteus M. H. Mammalian gene targeting with designed zinc finger nucleases. Mol. Ther. 13, 438–446 (2006). - PubMed
-
- Boch J. et al.. Breaking the code of DNA binding specificity of TAL-type III effectors. Science 326, 1509–1512 (2009). - PubMed
-
- Miller J. C. et al.. A TALE nuclease architecture for efficient genome editing. Nat. Biotechnol. 29, 143–148 (2011). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

