Sustaining Interferon Induction by a High-Passage Atypical Porcine Reproductive and Respiratory Syndrome Virus Strain
- PMID: 27805024
- PMCID: PMC5090871
- DOI: 10.1038/srep36312
Sustaining Interferon Induction by a High-Passage Atypical Porcine Reproductive and Respiratory Syndrome Virus Strain
Abstract
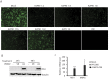
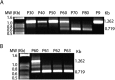
Porcine reproductive and respiratory syndrome virus (PRRSV) strain A2MC2 induces type I interferons in cultured cells. The objective of this study was to attenuate this strain by serial passaging in MARC-145 cells and assess its virulence and immunogenicity in pigs. The A2MC2 serially passaged 90 times (A2MC2-P90) retains the feature of interferon induction. The A2MC2-P90 replicates faster with a higher virus yield than wild type A2MC2 virus. Infection of primary pulmonary alveolar macrophages (PAMs) also induces interferons. Sequence analysis showed that the A2MC2-P90 has genomic nucleic acid identity of 99.8% to the wild type but has a deletion of 543 nucleotides in nsp2. The deletion occurred in passage 60. The A2MC2-P90 genome has a total of 35 nucleotide variations from the wild type, leading to 26 amino acid differences. Inoculation of three-week-old piglets showed that A2MC2-P90 is avirulent and elicits immune response. Compared with Ingelvac PRRS® MLV strain, A2MC2-P90 elicits higher virus neutralizing antibodies. The attenuated IFN-inducing A2MC2-P90 should be useful for development of an improved PRRSV vaccine.
Figures


References
-
- Holtkamp D. J. et al. Assessment of the economic impact of porcine reproductive and respiratory syndrome virus on U.S. pork producers. J Swine Health Prod. 21, 72–84 (2013).
-
- Meulenberg J. J. PRRSV, the virus. Vet Res. 31, 11–21 (2000). - PubMed
-
- Faaberg K. S. et al. Family Arteriviridae, in Virus taxonomy: classification and nomenclature of viruses: Ninth Report of the International Committee on Taxonomy of Viruses (eds King A. M. Q., Adams M. J., Carstens E. B. & Lefkowitz E. J.) (Elsevier Academic Press, San Diego, 2012).
-
- Rossow K. D. et al. Pathogenesis of porcine reproductive and respiratory syndrome virus infection in gnotobiotic pigs. Vet Pathol. 32, 361–373 (1995). - PubMed
-
- Kim H. S., Kwang J., Yoon I. J., Joo H. S. & Frey M. L. Enhanced replication of porcine reproductive and respiratory syndrome (PRRS) virus in a homogeneous subpopulation of MA-104 cell line. Arch Virol. 133, 477–483 (1993). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

