The Nucleic Acid Database: Present and Future
- PMID: 27805162
- PMCID: PMC4963143
- DOI: 10.6028/jres.101.026
The Nucleic Acid Database: Present and Future
Abstract
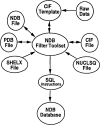
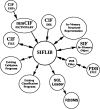
The Nucleic Acid Database is a relational database containing information about three-dimensional nucleic acid structures. The methods used for data processing, structure validation, database management and information retrieval, as well as the various services available via the World Wide Web, are described. Plans for the future include greater reliance on the Macromolecular Crystallographic Information File for both data processing and data management.
Keywords: Macro-molecular Crystallographic Information File; database management; nucleic acid structure.
Figures










Similar articles
-
PDB explorer -- a web based algorithm for protein annotation viewer and 3D visualization.Interdiscip Sci. 2014 Dec;6(4):279-84. doi: 10.1007/s12539-012-0044-x. Epub 2014 Aug 9. Interdiscip Sci. 2014. PMID: 25118648
-
NALDB: nucleic acid ligand database for small molecules targeting nucleic acid.Database (Oxford). 2016 Feb 20;2016:baw002. doi: 10.1093/database/baw002. Print 2016. Database (Oxford). 2016. PMID: 26896846 Free PMC article.
-
A comparative view at comprehensive information resources on three-dimensional structures of biological macro-molecules.Brief Funct Genomic Proteomic. 2007 Sep;6(3):220-39. doi: 10.1093/bfgp/elm020. Epub 2007 Oct 23. Brief Funct Genomic Proteomic. 2007. PMID: 17956938
-
Relational database design.J Insur Med. 2000;32(2):63-70. J Insur Med. 2000. PMID: 15912904 Review.
-
LinkHub: a Semantic Web system that facilitates cross-database queries and information retrieval in proteomics.BMC Bioinformatics. 2007 May 9;8 Suppl 3(Suppl 3):S5. doi: 10.1186/1471-2105-8-S3-S5. BMC Bioinformatics. 2007. PMID: 17493288 Free PMC article. Review.
Cited by
-
Incorporating Sequence-Dependent DNA Shape and Dynamics into Transcriptome Data Analysis.Methods Mol Biol. 2024;2812:317-343. doi: 10.1007/978-1-0716-3886-6_18. Methods Mol Biol. 2024. PMID: 39068371
References
-
- Bernstein EC, Koetzle TF, Williams GJB, Meyer EF, Brice MD, Rodgers JR, Kennard O, Shimanouchi T, Tasumi M. Protein Data Bank: A Computer-Based Archival File for Macromolecular Structures. J Mol Biol. 1977;112:535–542. - PubMed
-
- Macskassay S. Ndbview A Specialized 3-Dimensional Display Program for Crystallographic Structures of Nucleic Acids. Rutgers University; New Brunswick: 1991.
-
- Allen FH, Bellard S, Brice MD, Cartwright BA, Doubleday A, Higgs H, Hunnelink T, Kennard O, Motherwell WDS, Rogers JR, Watson DG. The Cambridge Crystallographic Data Centre: Computer-Based Search, Retrieval, Analysis and Display of Information. Acta Cryst. 1979;B35:2331–2339.
-
- Clowney L, Jain SC, Srinivasan AR, Westbrook J, Olson WK, Berman HM. Geometric Parameters In Nucleic Acids: Nitrogenous Bases. J Am Chem Soc. 1996;118:519–529.
LinkOut - more resources
Full Text Sources
