Forward-genetics analysis of sleep in randomly mutagenized mice
- PMID: 27806374
- PMCID: PMC6076225
- DOI: 10.1038/nature20142
Forward-genetics analysis of sleep in randomly mutagenized mice
Abstract
Sleep is conserved from invertebrates to vertebrates, and is tightly regulated in a homeostatic manner. The molecular and cellular mechanisms that determine the amount of rapid eye movement sleep (REMS) and non-REMS (NREMS) remain unknown. Here we identify two dominant mutations that affect sleep and wakefulness by using an electroencephalogram/electromyogram-based screen of randomly mutagenized mice. A splicing mutation in the Sik3 protein kinase gene causes a profound decrease in total wake time, owing to an increase in inherent sleep need. Sleep deprivation affects phosphorylation of regulatory sites on the kinase, suggesting a role for SIK3 in the homeostatic regulation of sleep amount. Sik3 orthologues also regulate sleep in fruitflies and roundworms. A missense, gain-of-function mutation in the sodium leak channel NALCN reduces the total amount and episode duration of REMS, apparently by increasing the excitability of REMS-inhibiting neurons. Our results substantiate the use of a forward-genetics approach for studying sleep behaviours in mice, and demonstrate the role of SIK3 and NALCN in regulating the amount of NREMS and REMS, respectively.
Conflict of interest statement
Author information
The authors declare no competing financial interests.
Figures
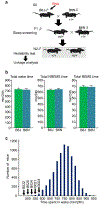

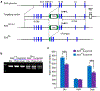

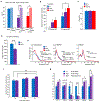










Comment in
-
Neuroscience: Sleepy and dreamless mutant mice.Nature. 2016 Nov 17;539(7629):364-365. doi: 10.1038/nature20471. Epub 2016 Nov 2. Nature. 2016. PMID: 27806380 No abstract available.
References
-
- Cirelli C et al. Reduced sleep in Drosophila Shaker mutants. Nature 434, 1087–1092 (2005). - PubMed
-
- Raizen DM et al. Lethargus is a Caenorhabditis elegans sleep-like state. Nature 451, 569–572 (2008). - PubMed
-
- Daan S, Beersma DG & Borbely, a a. Timing of human sleep: recovery process gated by a circadian pacemaker. Am. J. Physiol. 246, R161–83 (1984). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

