Developmental mechanisms of stripe patterns in rodents
- PMID: 27806375
- PMCID: PMC5292240
- DOI: 10.1038/nature20109
Developmental mechanisms of stripe patterns in rodents
Abstract
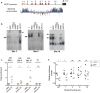
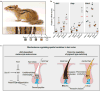
Mammalian colour patterns are among the most recognizable characteristics found in nature and can have a profound impact on fitness. However, little is known about the mechanisms underlying the formation and subsequent evolution of these patterns. Here we show that, in the African striped mouse (Rhabdomys pumilio), periodic dorsal stripes result from underlying differences in melanocyte maturation, which give rise to spatial variation in hair colour. We identify the transcription factor ALX3 as a regulator of this process. In embryonic dorsal skin, patterned expression of Alx3 precedes pigment stripes and acts to directly repress Mitf, a master regulator of melanocyte differentiation, thereby giving rise to light-coloured hair. Moreover, Alx3 is upregulated in the light stripes of chipmunks, which have independently evolved a similar dorsal pattern. Our results show a previously undescribed mechanism for modulating spatial variation in hair colour and provide insights into how phenotypic novelty evolves.
Figures
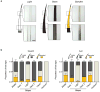
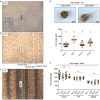

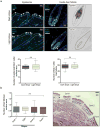

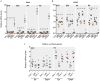

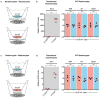
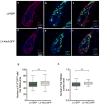


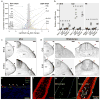

Comment in
-
Gnawing at striping - how rodents evolve striped patterns.Pigment Cell Melanoma Res. 2017 Mar;30(2):181-182. doi: 10.1111/pcmr.12580. Epub 2017 Mar 7. Pigment Cell Melanoma Res. 2017. PMID: 28182333 No abstract available.
References
-
- Barsh GS. The genetics of pigmentation: from fancy genes to complex traits. Trends in Genetics. 1996;12:299–305. - PubMed
-
- Jackson I, et al. Genetics and molecular biology of mouse pigmentation. Pigment Cell Research. 1994;7:73–80. - PubMed
-
- Rawls JF, Mellgren EM, Johnson SL. How the zebrafish gets its stripes. Developmental Biology. 2001;240:301–314. - PubMed
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

