DAAM1 stabilizes epithelial junctions by restraining WAVE complex-dependent lateral membrane motility
- PMID: 27807130
- PMCID: PMC5119936
- DOI: 10.1083/jcb.201603107
DAAM1 stabilizes epithelial junctions by restraining WAVE complex-dependent lateral membrane motility
Abstract
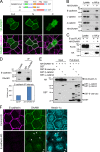
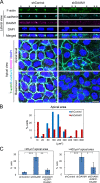
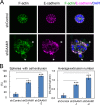
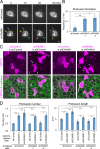
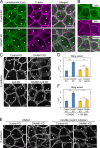
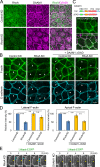
Epithelial junctions comprise two subdomains, the apical junctional complex (AJC) and the adjacent lateral membrane contacts (LCs), that span the majority of the junction. The AJC is lined with circumferential actin cables, whereas the LCs are associated with less-organized actin filaments whose roles are elusive. We found that DAAM1, a formin family actin regulator, accumulated at the LCs, and its depletion caused dispersion of actin filaments at these sites while hardly affecting circumferential actin cables. DAAM1 loss enhanced the motility of LC-forming membranes, leading to their invasion of neighboring cell layers, as well as disruption of polarized epithelial layers. We found that components of the WAVE complex and its downstream targets were required for the elevation of LC motility caused by DAAM1 loss. These findings suggest that the LC membranes are motile by nature because of the WAVE complex, but DAAM1-mediated actin regulation normally restrains this motility, thereby stabilizing epithelial architecture, and that DAAM1 loss evokes invasive abilities of epithelial cells.
© 2016 Nishimura et al.
Figures



References
-
- Carmona G., Perera U., Gillett C., Naba A., Law A.L., Sharma V.P., Wang J., Wyckoff J., Balsamo M., Mosis F., et al. . 2016. Lamellipodin promotes invasive 3D cancer cell migration via regulated interactions with Ena/VASP and SCAR/WAVE. Oncogene. 35:5155–5169. 10.1038/onc.2016.47 - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

