Asymmetrically localized proteins stabilize basal bodies against ciliary beating forces
- PMID: 27807131
- PMCID: PMC5119938
- DOI: 10.1083/jcb.201604135
Asymmetrically localized proteins stabilize basal bodies against ciliary beating forces
Abstract
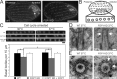
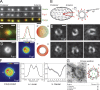
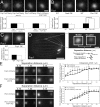
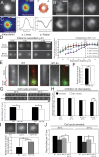
Basal bodies are radially symmetric, microtubule-rich structures that nucleate and anchor motile cilia. Ciliary beating produces asymmetric mechanical forces that are resisted by basal bodies. To resist these forces, distinct regions within the basal body ultrastructure and the microtubules themselves must be stable. However, the molecular components that stabilize basal bodies remain poorly defined. Here, we determine that Fop1 functionally interacts with the established basal body stability components Bld10 and Poc1. We find that Fop1 and microtubule glutamylation incorporate into basal bodies at distinct stages of assembly, culminating in their asymmetric enrichment at specific triplet microtubule regions that are predicted to experience the greatest mechanical force from ciliary beating. Both Fop1 and microtubule glutamylation are required to stabilize basal bodies against ciliary beating forces. Our studies reveal that microtubule glutamylation and Bld10, Poc1, and Fop1 stabilize basal bodies against the forces produced by ciliary beating via distinct yet interdependent mechanisms.
© 2016 Bayless et al.
Figures

References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

