Molecular Architecture of the Cleavage-Dependent Mannose Patch on a Soluble HIV-1 Envelope Glycoprotein Trimer
- PMID: 27807235
- PMCID: PMC5215339
- DOI: 10.1128/JVI.01894-16
Molecular Architecture of the Cleavage-Dependent Mannose Patch on a Soluble HIV-1 Envelope Glycoprotein Trimer
Abstract
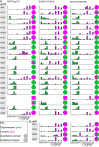
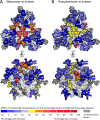
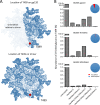
The formation of a correctly folded and natively glycosylated HIV-1 viral spike is dependent on protease cleavage of the gp160 precursor protein in the Golgi apparatus. Cleavage induces a compact structure which not only renders the spike capable of fusion but also limits further maturation of its extensive glycosylation. The redirection of the glycosylation pathway to preserve underprocessed oligomannose-type glycans is an important feature in immunogen design, as glycans contribute to or influence the epitopes of numerous broadly neutralizing antibodies. Here we present a quantitative site-specific analysis of a recombinant, trimeric mimic of the native HIV-1 viral spike (BG505 SOSIP.664) compared to the corresponding uncleaved pseudotrimer and the matched gp120 monomer. We present a detailed molecular map of a trimer-associated glycan remodeling that forms a localized subdomain of the native mannose patch. The formation of native trimers is a critical design feature in shaping the glycan epitopes presented on recombinant vaccine candidates.
Importance: The envelope spike of human immunodeficiency virus type 1 (HIV-1) is a target for antibody-based neutralization. For some patients infected with HIV-1, highly potent antibodies have been isolated that can neutralize a wide range of circulating viruses. It is a goal of HIV-1 vaccine research to elicit these antibodies by immunization with recombinant mimics of the viral spike. These antibodies have evolved to recognize the dense array of glycans that coat the surface of the viral molecule. We show how the structure of these glycans is shaped by steric constraints imposed upon them by the native folding of the viral spike. This information is important in guiding the development of vaccine candidates.
Keywords: furin; glycan; glycosylation; human immunodeficiency virus; neutralizing antibodies; oligosaccharides; structure; vaccines.
Copyright © 2017 American Society for Microbiology.
Figures



References
-
- Zhang M, Gaschen B, Blay W, Foley B, Haigwood N, Kuiken C, Korber B. 2004. Tracking global patterns of N-linked glycosylation site variation in highly variable viral glycoproteins: HIV, SIV, and HCV envelopes and influenza hemagglutinin. Glycobiology 14:1229–1246. doi:10.1093/glycob/cwh106. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

