T cell receptor recognition of CD1b presenting a mycobacterial glycolipid
- PMID: 27807341
- PMCID: PMC5095289
- DOI: 10.1038/ncomms13257
T cell receptor recognition of CD1b presenting a mycobacterial glycolipid
Abstract
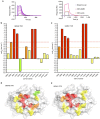
CD1 proteins present microbial lipids to T cells. Germline-encoded mycolyl lipid-reactive (GEM) T cells with conserved αβ T cell receptors (TCRs) recognize CD1b presenting mycobacterial mycolates. As the molecular basis underpinning TCR recognition of CD1b remains unknown, here we determine the structure of a GEM TCR bound to CD1b presenting glucose-6-O-monomycolate (GMM). The GEM TCR docks centrally above CD1b, whereby the conserved TCR α-chain extensively contacts CD1b and GMM. Through mutagenesis and study of T cells from tuberculosis patients, we identify a consensus CD1b footprint of TCRs present among GEM T cells. Using both the TCR α- and β-chains as tweezers to surround and grip the glucose moiety of GMM, GEM TCRs create a highly specific mechanism for recognizing this mycobacterial glycolipid.
Figures

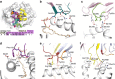

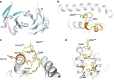

References
-
- Rossjohn J. et al.. T cell antigen receptor recognition of antigen-presenting molecules. Annu. Rev. Immunol. 33, 169–200 (2015). - PubMed
-
- Godfrey D. I., Uldrich A. P., McCluskey J., Rossjohn J. & Moody D. B. The burgeoning family of unconventional T cells. Nat. Immunol. 16, 1114–1123 (2015). - PubMed
-
- Salio M., Silk J. D., Jones E. Y. & Cerundolo V. Biology of CD1- and MR1-restricted T cells. Annu. Rev. Immunol. 32, 323–366 (2014). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

