Resistance to Biocides in Listeria monocytogenes Collected in Meat-Processing Environments
- PMID: 27807430
- PMCID: PMC5069283
- DOI: 10.3389/fmicb.2016.01627
Resistance to Biocides in Listeria monocytogenes Collected in Meat-Processing Environments
Abstract
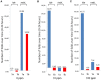
The emergence of microorganisms exerting resistance to biocides is a challenge to meat-processing environments. Bacteria can be intrinsically resistant to biocides but resistance can also be acquired by adaptation to their sub-lethal concentrations. Moreover, the presence of biocide resistance determinants, which is closely linked to antibiotic resistance determinants, could lead to co-selection during disinfection practices along the food chain, and select cross-resistant foodborne pathogens. The purpose of this work was to test the resistance of wild strains of Listeria monocytogenes, isolated from pork meat processing plants, toward benzalkonium chloride (BC), used as proxy of quaternary ammonium compounds. Furthermore, the expression of two non-specific efflux pumps genes (lde and mdrL) under biocide exposure was evaluated. L. monocytogenes were isolated from five processing plants located in the Veneto region (northeast of Italy) before and after cleaning and disinfection (C&D) procedures. A total of 45 strains were collected: 36 strains before and nine after the C&D procedures. Collected strains were typed according to MLST and ERIC profiles. Strains sampled in the same site, isolated before, and after the C&D procedures and displaying the same MLST and ERIC profiles were tested for their sensitivity to different concentrations of BC, in a time course assay. The expression of non-specific efflux pumps was evaluated at each time point by qPCR using tufA gene as housekeeping. A differential expression of the two investigated genes was observed: lde was found to be more expressed by the strains isolated before C&D procedures while its expression was dose-dependent in the case of the post C&D procedures strain. On the contrary, the expression of mdrL was inhibited under low biocidal stress (10 ppm BC) and enhanced in the presence of high stress (100 ppm BC). These findings suggests a possible role for C&D procedures to select L. monocytogenes persisters, pointing out the importance of dealing with the identification of risk factors in food plants sanification procedures that might select more tolerant strains.
Keywords: Listeria monocytogenes; efflux pumps; environment; food-processing plants; gene expression; resistance genes.
Figures

References
-
- Condell O., Iversen C., Cooney S., Power K. A., Walsh C., Burgess C., et al. (2012). Efficacy of biocides used in the modern food industry to control Salmonella enterica, and links between biocide tolerance and resistance to clinically relevant antimicrobial compounds. Appl. Environ. Microbiol. 78, 3087–3097. 10.1128/A.E.M.07534-11 - DOI - PMC - PubMed
-
- EU Commission (2004). Regulation (EC) No 852/2004. Off. J. Eur. Commun. 2002, 1–54.
LinkOut - more resources
Full Text Sources
Other Literature Sources

