Text Mining for Precision Medicine: Bringing Structure to EHRs and Biomedical Literature to Understand Genes and Health
- PMID: 27807747
- PMCID: PMC5931382
- DOI: 10.1007/978-981-10-1503-8_7
Text Mining for Precision Medicine: Bringing Structure to EHRs and Biomedical Literature to Understand Genes and Health
Abstract
The key question of precision medicine is whether it is possible to find clinically actionable granularity in diagnosing disease and classifying patient risk. The advent of next-generation sequencing and the widespread adoption of electronic health records (EHRs) have provided clinicians and researchers a wealth of data and made possible the precise characterization of individual patient genotypes and phenotypes. Unstructured text-found in biomedical publications and clinical notes-is an important component of genotype and phenotype knowledge. Publications in the biomedical literature provide essential information for interpreting genetic data. Likewise, clinical notes contain the richest source of phenotype information in EHRs. Text mining can render these texts computationally accessible and support information extraction and hypothesis generation. This chapter reviews the mechanics of text mining in precision medicine and discusses several specific use cases, including database curation for personalized cancer medicine, patient outcome prediction from EHR-derived cohorts, and pharmacogenomic research. Taken as a whole, these use cases demonstrate how text mining enables effective utilization of existing knowledge sources and thus promotes increased value for patients and healthcare systems. Text mining is an indispensable tool for translating genotype-phenotype data into effective clinical care that will undoubtedly play an important role in the eventual realization of precision medicine.
Keywords: Biomedical literature; Cancer; Database curation; EHR; Genotype; NLP; Outcome prediction; Pharmacogenomics; Phenotype; Precision medicine; Text mining.
Figures
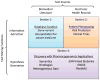



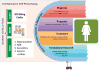

References
-
- Swede H, Stone CL, Norwood AR. National population-based biobanks for genetic research. Genet Med. 2007;9:141–149. - PubMed
-
- Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. - PubMed
-
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. - PubMed
-
- Garraway LA, Verweij J, Ballman KV. Precision oncology: an overview. J Clin Oncol. 2013;31:1803–1805. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

