DNA looping mediates nucleosome transfer
- PMID: 27808093
- PMCID: PMC5097161
- DOI: 10.1038/ncomms13337
DNA looping mediates nucleosome transfer
Abstract
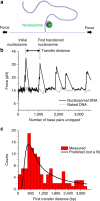
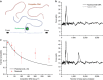
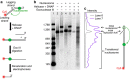
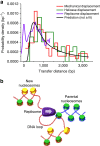
Proper cell function requires preservation of the spatial organization of chromatin modifications. Maintenance of this epigenetic landscape necessitates the transfer of parental nucleosomes to newly replicated DNA, a process that is stringently regulated and intrinsically linked to replication fork dynamics. This creates a formidable setting from which to isolate the central mechanism of transfer. Here we utilized a minimal experimental system to track the fate of a single nucleosome following its displacement, and examined whether DNA mechanics itself, in the absence of any chaperones or assembly factors, may serve as a platform for the transfer process. We found that the nucleosome is passively transferred to available dsDNA as predicted by a simple physical model of DNA loop formation. These results demonstrate a fundamental role for DNA mechanics in mediating nucleosome transfer and preserving epigenetic integrity during replication.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

