HnRNP L is important for the expression of oncogene SRSF3 and oncogenic potential of oral squamous cell carcinoma cells
- PMID: 27808105
- PMCID: PMC5093901
- DOI: 10.1038/srep35976
HnRNP L is important for the expression of oncogene SRSF3 and oncogenic potential of oral squamous cell carcinoma cells
Abstract
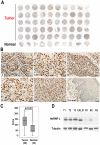
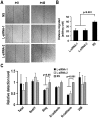
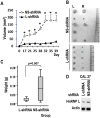
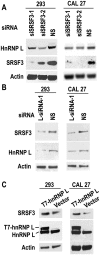
Oral squamous cell carcinoma (OSCC) is the leading cause of death related to oral diseases. The mechanisms of OSCC development remain largely unknown. Heterogeneous nuclear ribonucleoprotein L (HnRNP L) is a multi-functional splicing factor. It has been reported to be an important regulator of apoptosis. However, the functions of hnRNP L in cancer need to be further explored. In the present study, we found that OSCC tissues expressed significantly higher levels of hnRNP L than normal tissues. Depletion of hnRNP L retarded cell growth, cell migration, and tumorigenesis of OSCC cells. HnRNP L regulates both the expression of oncogenic splicing factor SRSF3 and the alternative splicing of SRSF3 exon 4. Expression of hnRNP L is correlated with SRSF3 expression in OSCC tissues. These findings suggest that hnRNP L is important for the pathogenesis of OSCC and may be a novel potential therapeutic target of OSCC.
Figures


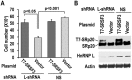

Similar articles
-
Inclusion of hnRNP L Alternative Exon 7 Is Associated with Good Prognosis and Inhibited by Oncogene SRSF3 in Head and Neck Squamous Cell Carcinoma.Biomed Res Int. 2019 Nov 13;2019:9612425. doi: 10.1155/2019/9612425. eCollection 2019. Biomed Res Int. 2019. PMID: 31828152 Free PMC article.
-
SRSF5 functions as a novel oncogenic splicing factor and is upregulated by oncogene SRSF3 in oral squamous cell carcinoma.Biochim Biophys Acta Mol Cell Res. 2018 Sep;1865(9):1161-1172. doi: 10.1016/j.bbamcr.2018.05.017. Epub 2018 May 30. Biochim Biophys Acta Mol Cell Res. 2018. PMID: 29857020
-
Expression of SRSF3 is Correlated with Carcinogenesis and Progression of Oral Squamous Cell Carcinoma.Int J Med Sci. 2016 Jun 30;13(7):533-9. doi: 10.7150/ijms.14871. eCollection 2016. Int J Med Sci. 2016. PMID: 27429590 Free PMC article.
-
Roles of FGFR in oral carcinogenesis.Cell Prolif. 2016 Jun;49(3):261-9. doi: 10.1111/cpr.12260. Cell Prolif. 2016. PMID: 27218663 Free PMC article. Review.
-
Oncogenic SRSF3 in health and diseases.Int J Biol Sci. 2023 Jun 12;19(10):3057-3076. doi: 10.7150/ijbs.83368. eCollection 2023. Int J Biol Sci. 2023. PMID: 37416784 Free PMC article. Review.
Cited by
-
Emerging roles of alternative RNA splicing in oral squamous cell carcinoma.Front Oncol. 2022 Nov 25;12:1019750. doi: 10.3389/fonc.2022.1019750. eCollection 2022. Front Oncol. 2022. PMID: 36505770 Free PMC article. Review.
-
SRSF3 functions as an oncogene in colorectal cancer by regulating the expression of ArhGAP30.Cancer Cell Int. 2020 Apr 10;20:120. doi: 10.1186/s12935-020-01201-2. eCollection 2020. Cancer Cell Int. 2020. PMID: 32308565 Free PMC article.
-
Biological function and molecular mechanism of SRSF3 in cancer and beyond.Oncol Lett. 2022 Jan;23(1):21. doi: 10.3892/ol.2021.13139. Epub 2021 Nov 16. Oncol Lett. 2022. PMID: 34858525 Free PMC article. Review.
-
Alternative RNA Splicing-The Trojan Horse of Cancer Cells in Chemotherapy.Genes (Basel). 2021 Jul 18;12(7):1085. doi: 10.3390/genes12071085. Genes (Basel). 2021. PMID: 34356101 Free PMC article. Review.
-
The role of alternative pre-mRNA splicing in cancer progression.Cancer Cell Int. 2023 Oct 24;23(1):249. doi: 10.1186/s12935-023-03094-3. Cancer Cell Int. 2023. PMID: 37875914 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

