Integrative omics connects N-glycoproteome-wide alterations with pathways and regulatory events in induced pluripotent stem cells
- PMID: 27808266
- PMCID: PMC5093713
- DOI: 10.1038/srep36109
Integrative omics connects N-glycoproteome-wide alterations with pathways and regulatory events in induced pluripotent stem cells
Abstract
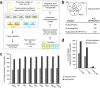
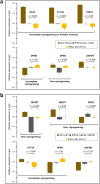
Molecular-level differences ranging from genomes to proteomes, but not N-glycoproteomes, between human induced pluripotent stem cells (hiPSCs) and embryonic stem cells (hESCs) have been assessed to gain insights into cell reprogramming and induced pluripotency. Our multiplexed quantitative N-glycoproteomics study identified altered N-glycoproteins that significantly regulate cell adhesion processes in hiPSCs compared to hESCs. The integrative proteomics and functional network analyses of the altered N-glycoproteins revealed their significant interactions with known PluriNet (pluripotency-associated network) proteins. We found that these interactions potentially regulate various signaling pathways including focal adhesion, PI3K-Akt signaling, regulation of actin cytoskeleton, and spliceosome. Furthermore, the integrative transcriptomics analysis revealed that imperfectly reprogrammed subunits of the oligosaccharyltransferase (OST) and dolichol-phosphate-mannose synthase (DPM) complexes were potential candidate regulatory events for the altered N-glycoprotein levels. Together, the results of our study suggest that imperfect reprogramming of the protein complexes linked with the N-glycosylation process may result in N-glycoprotein alterations that affect induced pluripotency through their functional protein interactions.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

