Post-transcriptional gene regulation by mRNA modifications
- PMID: 27808276
- PMCID: PMC5167638
- DOI: 10.1038/nrm.2016.132
Post-transcriptional gene regulation by mRNA modifications
Erratum in
-
Publisher Correction: Post-transcriptional gene regulation by mRNA modifications.Nat Rev Mol Cell Biol. 2018 Dec;19(12):808. doi: 10.1038/s41580-018-0075-1. Nat Rev Mol Cell Biol. 2018. PMID: 30341428
Abstract
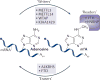
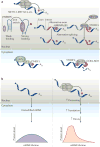
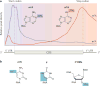
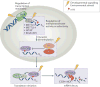
The recent discovery of reversible mRNA methylation has opened a new realm of post-transcriptional gene regulation in eukaryotes. The identification and functional characterization of proteins that specifically recognize RNA N6-methyladenosine (m6A) unveiled it as a modification that cells utilize to accelerate mRNA metabolism and translation. N6-adenosine methylation directs mRNAs to distinct fates by grouping them for differential processing, translation and decay in processes such as cell differentiation, embryonic development and stress responses. Other mRNA modifications, including N1-methyladenosine (m1A), 5-methylcytosine (m5C) and pseudouridine, together with m6A form the epitranscriptome and collectively code a new layer of information that controls protein synthesis.
Conflict of interest statement
statement The authors declare no competing interests.
Figures

References
-
- Perry RP, Kelley DE. Existence of methylated messenger RNA in mouse L cells. Cell. 1974;1:37–42. References 2 and 3 present the first reports of the existence of m6A in the internal region of mRNA.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

