Myotube differentiation in clustered regularly interspaced short palindromic repeat/Cas9-mediated MyoD knockout quail myoblast cells
- PMID: 27809462
- PMCID: PMC5495663
- DOI: 10.5713/ajas.16.0749
Myotube differentiation in clustered regularly interspaced short palindromic repeat/Cas9-mediated MyoD knockout quail myoblast cells
Abstract
Objective: In the livestock industry, the regulatory mechanisms of muscle proliferation and differentiation can be applied to improve traits such as growth and meat production. We investigated the regulatory pathway of MyoD and its role in muscle differentiation in quail myoblast cells.
Methods: The MyoD gene was mutated by the clustered regularly interspaced short palindromic repeat (CRISPR)/Cas9 technology and single cell-derived MyoD mutant sublines were identified to investigate the global regulatory mechanism responsible for muscle differentiation.
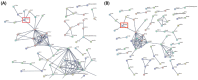
Results: The mutation efficiency was 73.3% in the mixed population, and from this population we were able to establish two QM7 MyoD knockout subline (MyoD KO QM7#4) through single cell pick-up and expansion. In the undifferentiated condition, paired box 7 expression in MyoD KO QM7#4 cells was not significantly different from regular QM7 (rQM7) cells. During differentiation, however, myotube formation was dramatically repressed in MyoD KO QM7#4 cells. Moreover, myogenic differentiation-specific transcripts and proteins were not expressed in MyoD KO QM7#4 cells even after an extended differentiation period. These results indicate that MyoD is critical for muscle differentiation. Furthermore, we analyzed the global regulatory interactions by RNA sequencing during muscle differentiation.
Conclusion: With CRISPR/Cas9-mediated genomic editing, single cell-derived sublines with a specific knockout gene can be adapted to various aspects of basic research as well as in functional genomics studies.
Keywords: CRISPR-Cas9; Knockout; Muscle Differentiation; MyoD; Myoblast.
Conflict of interest statement
We certify that there is no conflict of interest with any financial organization regarding the material discussed in the manuscript.
Figures




References
-
- Hillier LW, Miller W, Birney E, et al. Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature. 2004;432:695–716. - PubMed
-
- Rubin CJ, Zody MC, Eriksson J, et al. Whole-genome resequencing reveals loci under selection during chicken domestication. Nature. 2010;464:587–91. - PubMed
-
- Shin S, Song Y, Ahn J, et al. A novel mechanism of myostatin regulation by its alternative splicing variant during myogenesis in avian species. Am J Physiol Cell Physiol. 2015;309:650–9. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

