Patient-Specific and Genome-Edited Induced Pluripotent Stem Cell-Derived Cardiomyocytes Elucidate Single-Cell Phenotype of Brugada Syndrome
- PMID: 27810048
- PMCID: PMC5373649
- DOI: 10.1016/j.jacc.2016.07.779
Patient-Specific and Genome-Edited Induced Pluripotent Stem Cell-Derived Cardiomyocytes Elucidate Single-Cell Phenotype of Brugada Syndrome
Abstract
Background: Brugada syndrome (BrS), a disorder associated with characteristic electrocardiogram precordial ST-segment elevation, predisposes afflicted patients to ventricular fibrillation and sudden cardiac death. Despite marked achievements in outlining the organ level pathophysiology of the disorder, the understanding of human cellular phenotype has lagged due to a lack of adequate human cellular models of the disorder.
Objectives: The objective of this study was to examine single cell mechanism of Brugada syndrome using induced pluripotent stem cell-derived cardiomyocytes (iPSC-CMs).
Methods: This study recruited 2 patients with type 1 BrS carrying 2 different sodium voltage-gated channel alpha subunit 5 variants as well as 2 healthy control subjects. We generated iPSCs from their skin fibroblasts by using integration-free Sendai virus. We used directed differentiation to create purified populations of iPSC-CMs.
Results: BrS iPSC-CMs showed reductions in inward sodium current density and reduced maximal upstroke velocity of action potential compared with healthy control iPSC-CMs. Furthermore, BrS iPSC-CMs demonstrated increased burden of triggered activity, abnormal calcium (Ca2+) transients, and beating interval variation. Correction of the causative variant by genome editing was performed, and resultant iPSC-CMs showed resolution of triggered activity and abnormal Ca2+ transients. Gene expression profiling of iPSC-CMs showed clustering of BrS compared with control subjects. Furthermore, BrS iPSC-CM gene expression correlated with gene expression from BrS human cardiac tissue gene expression.
Conclusions: Patient-specific iPSC-CMs were able to recapitulate single-cell phenotype features of BrS, including blunted inward sodium current, increased triggered activity, and abnormal Ca2+ handling. This novel human cellular model creates future opportunities to further elucidate the cellular disease mechanism and identify novel therapeutic targets.
Keywords: Ca(2+) transient; SCN5A; action potential; arrhythmia; gene expression; genome editing.
Copyright © 2016 American College of Cardiology Foundation. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
The authors have no conflicts of interest to disclose.
Figures
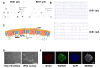


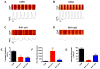
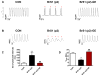
Comment in
-
Modeling and Genome-Editing Brugada Syndrome in a Dish.J Am Coll Cardiol. 2016 Nov 8;68(19):2097-2098. doi: 10.1016/j.jacc.2016.09.911. J Am Coll Cardiol. 2016. PMID: 27810049 Free PMC article. No abstract available.
References
-
- Brugada P, Brugada J. Right bundle branch block, persistent ST segment elevation and sudden cardiac death: a distinct clinical and electrocardiographic syndrome. A multicenter report. J Am Coll Cardiol. 1992;20:1391–6. - PubMed
-
- Brugada J, Brugada R, Brugada P. Determinants of sudden cardiac death in individuals with the electrocardiographic pattern of Brugada syndrome and no previous cardiac arrest. Circulation. 2003;108:3092–6. - PubMed
-
- Antzelevitch C, Brugada P, Borggrefe M, et al. Brugada syndrome: report of the second consensus conference: endorsed by the Heart Rhythm Society and the European Heart Rhythm Association. Circulation. 2005;111:659–70. - PubMed
-
- Veltmann C, Schimpf R, Echternach C, et al. A prospective study on spontaneous fluctuations between diagnostic and non-diagnostic ECGs in Brugada syndrome: implications for correct phenotyping and risk stratification. Eur Heart J. 2006;27:2544–52. - PubMed
-
- Yan GX, Antzelevitch C. Cellular basis for the Brugada syndrome and other mechanisms of arrhythmogenesis associated with ST-segment elevation. Circulation. 1999;100:1660–6. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

