CD72 negatively regulates B lymphocyte responses to the lupus-related endogenous toll-like receptor 7 ligand Sm/RNP
- PMID: 27810925
- PMCID: PMC5110020
- DOI: 10.1084/jem.20160560
CD72 negatively regulates B lymphocyte responses to the lupus-related endogenous toll-like receptor 7 ligand Sm/RNP
Abstract
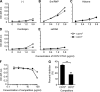
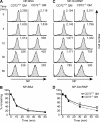
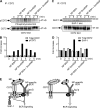
Toll-like receptor 7 (TLR7) plays an essential role in development of systemic lupus erythematosus by co-stimulating B cells reactive to the endogenous TLR7 ligand Sm/ribonucleoprotein (RNP), a crucial lupus self-antigen. However, how the TLR7-mediated autoimmune response is regulated is not yet known. In this study, we demonstrate that CD72, an inhibitory B cell co-receptor known to prevent development of lupus, recognizes Sm/RNP at the extracellular C-type lectin-like domain (CTLD) and specifically inhibits B cell response to Sm/RNP. Moreover, the CTLD of CD72c, a lupus-susceptible allele, binds to Sm/RNP less strongly than that of lupus-resistant CD72a Reduced binding of CD72c is supported by x-ray crystallographic analysis that reveals a considerable alteration in charge at the putative ligand-binding site. Thus, CD72 appears to specifically inhibit B cell response to the endogenous TLR7 ligand Sm/RNP through CTLD-mediated recognition of Sm/RNP, thereby preventing production of anti-Sm/RNP antibody crucial for development of lupus.
© 2016 Akatsu et al.
Figures





References
-
- Adachi T., Flaswinkel H., Yakura H., Reth M., and Tsubata T.. 1998. The B cell surface protein CD72 recruits the tyrosine phosphatase SHP-1 upon tyrosine phosphorylation. J. Immunol. 160:4662–4665. - PubMed
-
- Adams P.D., Afonine P.V., Bunkóczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W., et al. . 2010. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66:213–221. 10.1107/S0907444909052925 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

