The Protein Cost of Metabolic Fluxes: Prediction from Enzymatic Rate Laws and Cost Minimization
- PMID: 27812109
- PMCID: PMC5094713
- DOI: 10.1371/journal.pcbi.1005167
The Protein Cost of Metabolic Fluxes: Prediction from Enzymatic Rate Laws and Cost Minimization
Abstract
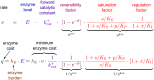
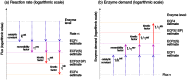
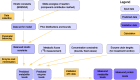
Bacterial growth depends crucially on metabolic fluxes, which are limited by the cell's capacity to maintain metabolic enzymes. The necessary enzyme amount per unit flux is a major determinant of metabolic strategies both in evolution and bioengineering. It depends on enzyme parameters (such as kcat and KM constants), but also on metabolite concentrations. Moreover, similar amounts of different enzymes might incur different costs for the cell, depending on enzyme-specific properties such as protein size and half-life. Here, we developed enzyme cost minimization (ECM), a scalable method for computing enzyme amounts that support a given metabolic flux at a minimal protein cost. The complex interplay of enzyme and metabolite concentrations, e.g. through thermodynamic driving forces and enzyme saturation, would make it hard to solve this optimization problem directly. By treating enzyme cost as a function of metabolite levels, we formulated ECM as a numerically tractable, convex optimization problem. Its tiered approach allows for building models at different levels of detail, depending on the amount of available data. Validating our method with measured metabolite and protein levels in E. coli central metabolism, we found typical prediction fold errors of 4.1 and 2.6, respectively, for the two kinds of data. This result from the cost-optimized metabolic state is significantly better than randomly sampled metabolite profiles, supporting the hypothesis that enzyme cost is important for the fitness of E. coli. ECM can be used to predict enzyme levels and protein cost in natural and engineered pathways, and could be a valuable computational tool to assist metabolic engineering projects. Furthermore, it establishes a direct connection between protein cost and thermodynamics, and provides a physically plausible and computationally tractable way to include enzyme kinetics into constraint-based metabolic models, where kinetics have usually been ignored or oversimplified.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
-
- Warburg O, Posener K, Negelein E. Ueber den Stoffwechsel der Tumoren. Biochemische Zeitschrift. 1924;152:319–344.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

