Tensor Analysis Reveals Distinct Population Structure that Parallels the Different Computational Roles of Areas M1 and V1
- PMID: 27814353
- PMCID: PMC5096707
- DOI: 10.1371/journal.pcbi.1005164
Tensor Analysis Reveals Distinct Population Structure that Parallels the Different Computational Roles of Areas M1 and V1
Abstract
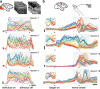
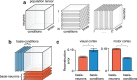
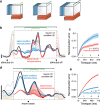
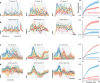
Cortical firing rates frequently display elaborate and heterogeneous temporal structure. One often wishes to compute quantitative summaries of such structure-a basic example is the frequency spectrum-and compare with model-based predictions. The advent of large-scale population recordings affords the opportunity to do so in new ways, with the hope of distinguishing between potential explanations for why responses vary with time. We introduce a method that assesses a basic but previously unexplored form of population-level structure: when data contain responses across multiple neurons, conditions, and times, they are naturally expressed as a third-order tensor. We examined tensor structure for multiple datasets from primary visual cortex (V1) and primary motor cortex (M1). All V1 datasets were 'simplest' (there were relatively few degrees of freedom) along the neuron mode, while all M1 datasets were simplest along the condition mode. These differences could not be inferred from surface-level response features. Formal considerations suggest why tensor structure might differ across modes. For idealized linear models, structure is simplest across the neuron mode when responses reflect external variables, and simplest across the condition mode when responses reflect population dynamics. This same pattern was present for existing models that seek to explain motor cortex responses. Critically, only dynamical models displayed tensor structure that agreed with the empirical M1 data. These results illustrate that tensor structure is a basic feature of the data. For M1 the tensor structure was compatible with only a subset of existing models.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
-
- Bair W, Koch C. Temporal precision of spike trains in extrastriate cortex of the behaving macaque monkey. Neural Comput. 1996. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

