Deleting an Nr4a1 Super-Enhancer Subdomain Ablates Ly6Clow Monocytes while Preserving Macrophage Gene Function
- PMID: 27814941
- PMCID: PMC5694686
- DOI: 10.1016/j.immuni.2016.10.011
Deleting an Nr4a1 Super-Enhancer Subdomain Ablates Ly6Clow Monocytes while Preserving Macrophage Gene Function
Abstract
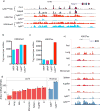
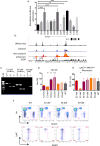
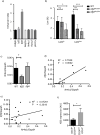
Mononuclear phagocytes are a heterogeneous family that occupy all tissues and assume numerous roles to support tissue function and systemic homeostasis. Our ability to dissect the roles of individual subsets is limited by a lack of technologies that ablate gene function within specific mononuclear phagocyte sub-populations. Using Nr4a1-dependent Ly6Clow monocytes, we present a proof-of-principle approach that addresses these limitations. Combining ChIP-seq and molecular approaches we identified a single, conserved, sub-domain within the Nr4a1 enhancer that was essential for Ly6Clow monocyte development. Mice lacking this enhancer lacked Ly6Clow monocytes but retained Nr4a1 gene expression in macrophages during steady state and in response to LPS. Because Nr4a1 regulates inflammatory gene expression and differentiation of Ly6Clow monocytes, decoupling these processes allows Ly6Clow monocytes to be studied independently.
Copyright © 2016 Elsevier Inc. All rights reserved.
Figures




Comment in
-
The Ly6C(lo)-Down: Targeting Developmental Enhancers for Cell Subset Depletion.Immunity. 2016 Nov 15;45(5):949-951. doi: 10.1016/j.immuni.2016.10.034. Immunity. 2016. PMID: 27851919
References
-
- Amit I, Winter DR, Jung S. The role of the local environment and epigenetics in shaping macrophage identity and their effect on tissue homeostasis. Nat Immunol. 2016;17:18–25. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P01 HL088093/HL/NHLBI NIH HHS/United States
- R01 GM086912/GM/NIGMS NIH HHS/United States
- R01 HL086548/HL/NHLBI NIH HHS/United States
- S10 RR027366/RR/NCRR NIH HHS/United States
- R01 DK091183/DK/NIDDK NIH HHS/United States
- P30 DK063491/DK/NIDDK NIH HHS/United States
- R01 CA202987/CA/NCI NIH HHS/United States
- R00 HL123485/HL/NHLBI NIH HHS/United States
- R01 HL118765/HL/NHLBI NIH HHS/United States
- P01 DK074868/DK/NIDDK NIH HHS/United States
- 12SDG12070005/AHA/American Heart Association-American Stroke Association/United States
- P30 CA023100/CA/NCI NIH HHS/United States
- T32 GM008666/GM/NIGMS NIH HHS/United States
- R01 HL134236/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

