High-level ROR1 associates with accelerated disease progression in chronic lymphocytic leukemia
- PMID: 27815263
- PMCID: PMC5179332
- DOI: 10.1182/blood-2016-04-712562
High-level ROR1 associates with accelerated disease progression in chronic lymphocytic leukemia
Abstract
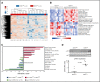
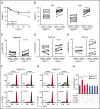
ROR1 is an oncoembryonic orphan receptor found on chronic lymphocytic leukemia (CLL) B cells, but not on normal postpartum tissues. ROR1 is a receptor for Wnt5a that may complex with TCL1, a coactivator of AKT that is able to promote development of CLL. We found the CLL cells of a few patients expressed negligible ROR1 (ROR1Neg), but expressed TCL1A at levels comparable to those of samples that expressed ROR1 (ROR1Pos). Transcriptome analyses revealed that ROR1Neg cases generally could be distinguished from those that were ROR1Pos in unsupervised gene-expression clustering analysis. Gene-set enrichment analyses demonstrated that ROR1Neg CLL had lower expression and activation of AKT signaling pathways relative to ROR1Pos CLL, similar to what was noted for leukemia that respectively developed in TCL1 vs ROR1xTCL1 transgenic mice. In contrast to its effect on ROR1Pos CLL, Wnt5a did not enhance the proliferation, chemotaxis, or survival of ROR1Neg CLL. We examined the CLL cells from 1568 patients, which we randomly assigned to a training or validation set of 797 or 771 cases, respectively. Using recursive partitioning, we defined a threshold for ROR1 surface expression that could segregate samples of the training set into ROR1-Hi vs ROR1-Lo subgroups that differed significantly in their median treatment-free survival (TFS). Using this threshold, we found that ROR1-Hi cases had a significantly shorter median TFS and overall survival than ROR1-Lo cases in the validation set. These data demonstrate that expression of ROR1 may promote leukemia-cell activation and survival and enhance disease progression in patients with CLL.
© 2016 by The American Society of Hematology.
Figures



References
-
- Baskar S, Kwong KY, Hofer T, et al. Unique cell surface expression of receptor tyrosine kinase ROR1 in human B-cell chronic lymphocytic leukemia. Clin Cancer Res. 2008;14(2):396-404. - PubMed
-
- Daneshmanesh AH, Mikaelsson E, Jeddi-Tehrani M, et al. Ror1, a cell surface receptor tyrosine kinase is expressed in chronic lymphocytic leukemia and may serve as a putative target for therapy. Int J Cancer. 2008;123(5):1190-1195. - PubMed
-
- Choudhury A, Derkow K, Daneshmanesh AH, et al. Silencing of ROR1 and FMOD with siRNA results in apoptosis of CLL cells. Br J Haematol. 2010;151(4):327-335. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

