Genetic markers associated with dihydroartemisinin-piperaquine failure in Plasmodium falciparum malaria in Cambodia: a genotype-phenotype association study
- PMID: 27818095
- PMCID: PMC5564489
- DOI: 10.1016/S1473-3099(16)30409-1
Genetic markers associated with dihydroartemisinin-piperaquine failure in Plasmodium falciparum malaria in Cambodia: a genotype-phenotype association study
Abstract
Background: As the prevalence of artemisinin-resistant Plasmodium falciparum malaria increases in the Greater Mekong subregion, emerging resistance to partner drugs in artemisinin combination therapies seriously threatens global efforts to treat and eliminate this disease. Molecular markers that predict failure of artemisinin combination therapy are urgently needed to monitor the spread of partner drug resistance, and to recommend alternative treatments in southeast Asia and beyond.
Methods: We did a genome-wide association study of 297 P falciparum isolates from Cambodia to investigate the relationship of 11 630 exonic single-nucleotide polymorphisms (SNPs) and 43 copy number variations (CNVs) with in-vitro piperaquine 50% inhibitory concentrations (IC50s), and tested whether these genetic variants are markers of treatment failure with dihydroartemisinin-piperaquine. We then did a survival analysis of 133 patients to determine whether candidate molecular markers predicted parasite recrudescence following dihydroartemisinin-piperaquine treatment.
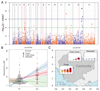
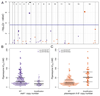
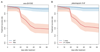
Findings: Piperaquine IC50s increased significantly from 2011 to 2013 in three Cambodian provinces (2011 vs 2013 median IC50s: 20·0 nmol/L [IQR 13·7-29·0] vs 39·2 nmol/L [32·8-48·1] for Ratanakiri, 19·3 nmol/L [15·1-26·2] vs 66·2 nmol/L [49·9-83·0] for Preah Vihear, and 19·6 nmol/L [11·9-33·9] vs 81·1 nmol/L [61·3-113·1] for Pursat; all p≤10-3; Kruskal-Wallis test). Genome-wide analysis of SNPs identified a chromosome 13 region that associates with raised piperaquine IC50s. A non-synonymous SNP (encoding a Glu415Gly substitution) in this region, within a gene encoding an exonuclease, associates with parasite recrudescence following dihydroartemisinin-piperaquine treatment. Genome-wide analysis of CNVs revealed that a single copy of the mdr1 gene on chromosome 5 and a novel amplification of the plasmepsin 2 and plasmepsin 3 genes on chromosome 14 also associate with raised piperaquine IC50s. After adjusting for covariates, both exo-E415G and plasmepsin 2-3 markers significantly associate (p=3·0 × 10-8 and p=1·7 × 10-7, respectively) with decreased treatment efficacy (survival rates 0·38 [95% CI 0·25-0·51] and 0·41 [0·28-0·53], respectively).
Interpretation: The exo-E415G SNP and plasmepsin 2-3 amplification are markers of piperaquine resistance and dihydroartemisinin-piperaquine failures in Cambodia, and can help monitor the spread of these phenotypes into other countries of the Greater Mekong subregion, and elucidate the mechanism of piperaquine resistance. Since plasmepsins are involved in the parasite's haemoglobin-to-haemozoin conversion pathway, targeted by related antimalarials, plasmepsin 2-3 amplification probably mediates piperaquine resistance.
Funding: Intramural Research Program of the US National Institute of Allergy and Infectious Diseases, National Institutes of Health, Wellcome Trust, Bill & Melinda Gates Foundation, Medical Research Council, and UK Department for International Development.
Copyright © 2017 The Author(s). Published by Elsevier Ltd. This is an Open Access article under the CC BY license. Published by Elsevier Ltd.. All rights reserved.
Conflict of interest statement
We declare that we have no conflicts of interest.
Figures
Comment in
-
New genetic marker for piperaquine resistance in Plasmodium falciparum.Lancet Infect Dis. 2017 Feb;17(2):119-121. doi: 10.1016/S1473-3099(16)30414-5. Epub 2016 Nov 3. Lancet Infect Dis. 2017. PMID: 27818096 No abstract available.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

