Removal of oxidatively generated DNA damage by overlapping repair pathways
- PMID: 27818219
- PMCID: PMC5418118
- DOI: 10.1016/j.freeradbiomed.2016.10.507
Removal of oxidatively generated DNA damage by overlapping repair pathways
Abstract
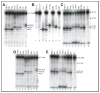
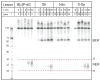
It is generally believed that the mammalian nucleotide excision repair pathway removes DNA helix-distorting bulky DNA lesions, while small non-bulky lesions are repaired by base excision repair (BER). However, recent work demonstrates that the oxidativly generated guanine oxidation products, spiroimininodihydantoin (Sp), 5-guanidinohydantoin (Gh), and certain intrastrand cross-linked lesions, are good substrates of NER and BER pathways that compete with one another in human cell extracts. The oxidation of guanine by peroxynitrite is known to generate 5-guanidino-4-nitroimidazole (NIm) which is structurally similar to Gh, except that the 4-nitro group in NIm is replaced by a keto group in Gh. However, unlike Gh, NIm is an excellent substrate of BER, but not of NER. These and other related results are reviewed and discussed in this article.
Keywords: Base excision repair; DNA damage; Nucleotide excision repair; Oxidative stress; Reactive oxygen species.
Copyright © 2017 Elsevier Inc. All rights reserved.
Figures
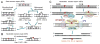

References
-
- Hussain SP, Harris CC. Inflammation and cancer: an ancient link with novel potentials. Int J Cancer. 2007;121:2373–2380. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

