Overexpression of a Barley Aquaporin Gene, HvPIP2;5 Confers Salt and Osmotic Stress Tolerance in Yeast and Plants
- PMID: 27818670
- PMCID: PMC5073208
- DOI: 10.3389/fpls.2016.01566
Overexpression of a Barley Aquaporin Gene, HvPIP2;5 Confers Salt and Osmotic Stress Tolerance in Yeast and Plants
Abstract
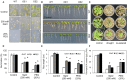
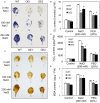
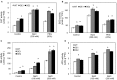
We characterized an aquaporin gene HvPIP2;5 from Hordeum vulgare and investigated its physiological roles in heterologous expression systems, yeast and Arabidopsis, under high salt and high osmotic stress conditions. In yeast, the expression of HvPIP2;5 enhanced abiotic stress tolerance under high salt and high osmotic conditions. Arabidopsis plants overexpressing HvPIP2;5 also showed better stress tolerance in germination and root growth under high salt and high osmotic stresses than the wild type (WT). HvPIP2;5 overexpressing plants were able to survive and recover after a 3-week drought period unlike the control plants which wilted and died during stress treatment. Indeed, overexpression of HvPIP2;5 caused higher retention of chlorophylls and water under salt and osmotic stresses than did control. We also observed lower accumulation of reactive oxygen species (ROS) and malondialdehyde (MDA), an end-product of lipid peroxidation in HvPIP2;5 overexpressing plants than in WT. These results suggest that HvPIP2;5 overexpression brought about stress tolerance, at least in part, by reducing the secondary oxidative stress caused by salt and osmotic stresses. Consistent with these stress tolerant phenotypes, HvPIP2;5 overexpressing Arabidopsis lines showed higher expression and activities of ROS scavenging enzymes such as catalase (CAT), superoxide dismutase (SOD), glutathione reductase (GR), and ascorbate peroxidase (APX) under salt and osmotic stresses than did WT. In addition, the proline biosynthesis genes, Δ 1-Pyrroline-5-Carboxylate Synthase 1 and 2 (P5CS1 and P5CS2) were up-regulated in HvPIP2;5 overexpressing plants under salt and osmotic stresses, which coincided with increased levels of the osmoprotectant proline. Together, these results suggested that HvPIP2;5 overexpression enhanced stress tolerance to high salt and high osmotic stresses by increasing activities and/or expression of ROS scavenging enzymes and osmoprotectant biosynthetic genes.
Keywords: Arabidopsis; HvPIP2;5; aquaporin; barley; overexpression; stress tolerance; yeast.
Figures


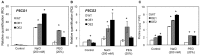
References
-
- Aharon R., Shahak Y., Wininger S., Bendov R., Kapulnik Y., Galili G. (2003). Overexpression of a plasma membrane aquaporin in transgenic tobacco improves plant vigor under favorable growth conditions but not under drought or salt stress. Plant Cell 15, 439–447. 10.1105/tpc.009225 - DOI - PMC - PubMed
-
- Bates L. S., Waldren R. P., Teare I. D. (1973). Rapid determination of free proline for water-stress studies. Plant Soil 39, 205–207. 10.1007/BF00018060 - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

