Stringent comparative sequence analysis reveals SOX10 as a putative inhibitor of glial cell differentiation
- PMID: 27821050
- PMCID: PMC5100263
- DOI: 10.1186/s12864-016-3167-3
Stringent comparative sequence analysis reveals SOX10 as a putative inhibitor of glial cell differentiation
Abstract
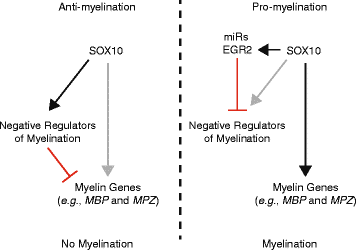
Background: The transcription factor SOX10 is essential for all stages of Schwann cell development including myelination. SOX10 cooperates with other transcription factors to activate the expression of key myelin genes in Schwann cells and is therefore a context-dependent, pro-myelination transcription factor. As such, the identification of genes regulated by SOX10 will provide insight into Schwann cell biology and related diseases. While genome-wide studies have successfully revealed SOX10 target genes, these efforts mainly focused on myelinating stages of Schwann cell development. We propose that less-biased approaches will reveal novel functions of SOX10 outside of myelination.
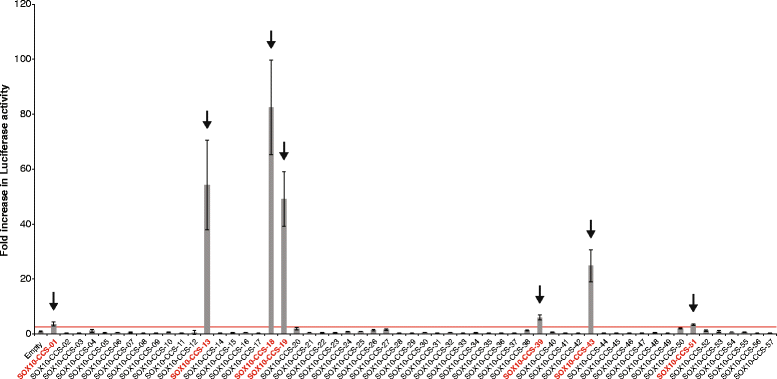
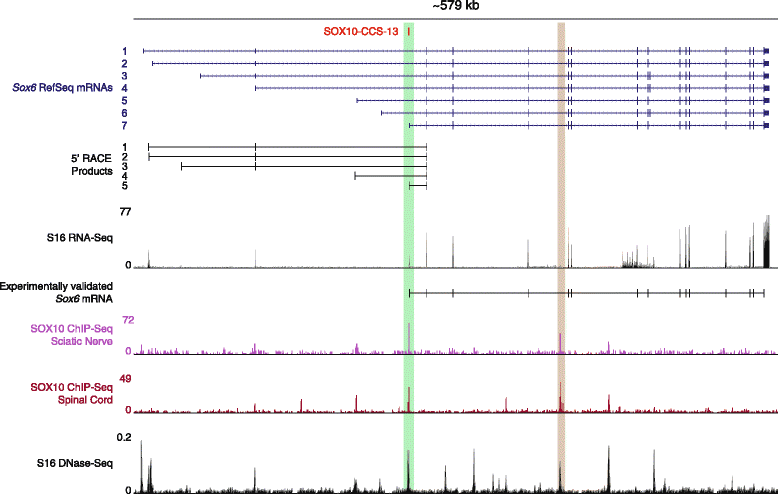
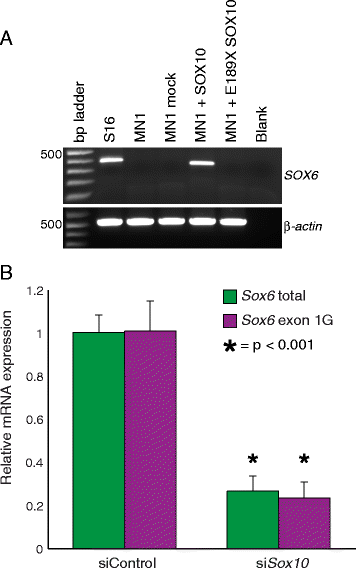
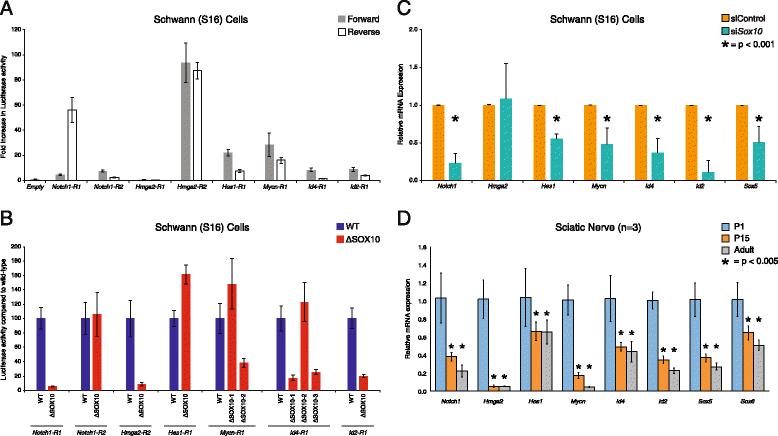
Results: We developed a stringent, computational-based screen for genome-wide identification of SOX10 response elements. Experimental validation of a pilot set of predicted binding sites in multiple systems revealed that SOX10 directly regulates a previously unreported alternative promoter at SOX6, which encodes a transcription factor that inhibits glial cell differentiation. We further explored the utility of our computational approach by combining it with DNase-seq analysis in cultured Schwann cells and previously published SOX10 ChIP-seq data from rat sciatic nerve. Remarkably, this analysis enriched for genomic segments that map to loci involved in the negative regulation of gliogenesis including SOX5, SOX6, NOTCH1, HMGA2, HES1, MYCN, ID4, and ID2. Functional studies in Schwann cells revealed that: (1) all eight loci are expressed prior to myelination and down-regulated subsequent to myelination; (2) seven of the eight loci harbor validated SOX10 binding sites; and (3) seven of the eight loci are down-regulated upon repressing SOX10 function.
Conclusions: Our computational strategy revealed a putative novel function for SOX10 in Schwann cells, which suggests a model where SOX10 activates the expression of genes that inhibit myelination during non-myelinating stages of Schwann cell development. Importantly, the computational and functional datasets we present here will be valuable for the study of transcriptional regulation, SOX protein function, and glial cell biology.
Keywords: Comparative sequence analysis; Myelination; SOX10; Schwann cells; Transcriptional regulation; Ultra-conserved sequences.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

