Role for the MED21-MED7 Hinge in Assembly of the Mediator-RNA Polymerase II Holoenzyme
- PMID: 27821593
- PMCID: PMC5207194
- DOI: 10.1074/jbc.M116.756098
Role for the MED21-MED7 Hinge in Assembly of the Mediator-RNA Polymerase II Holoenzyme
Abstract
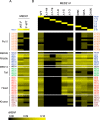
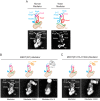
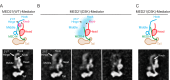
Mediator plays an integral role in activation of RNA polymerase II (Pol II) transcription. A key step in activation is binding of Mediator to Pol II to form the Mediator-Pol II holoenzyme. Here, we exploit a combination of biochemistry and macromolecular EM to investigate holoenzyme assembly. We identify a subset of human Mediator head module subunits that bind Pol II independent of other subunits and thus probably contribute to a major Pol II binding site. In addition, we show that binding of human Mediator to Pol II depends on the integrity of a conserved "hinge" in the middle module MED21-MED7 heterodimer. Point mutations in the hinge region leave core Mediator intact but lead to increased disorder of the middle module and markedly reduced affinity for Pol II. These findings highlight the importance of Mediator conformation for holoenzyme assembly.
Keywords: Mediator; RNA polymerase II; general transcription factor (GTF); transcription; transcription coregulator; transcription regulation.
© 2016 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures




References
-
- Kim Y. J., Björklund S., Li Y., Sayre M. H., and Kornberg R. D. (1994) A multiprotein mediator of transcriptional activation and its interaction with the C-terminal repeat domain of RNA polymerase II. Cell 77, 599–608 - PubMed
-
- Myers L. C., and Kornberg R. D. (2000) Mediator of Transcriptional Regulation. Annu. Rev. Biochem. 69, 729–749 - PubMed
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

