APOL1 Renal-Risk Variants Induce Mitochondrial Dysfunction
- PMID: 27821631
- PMCID: PMC5373457
- DOI: 10.1681/ASN.2016050567
APOL1 Renal-Risk Variants Induce Mitochondrial Dysfunction
Abstract
APOL1 G1 and G2 variants facilitate kidney disease in blacks. To elucidate the pathways whereby these variants contribute to disease pathogenesis, we established HEK293 cell lines stably expressing doxycycline-inducible (Tet-on) reference APOL1 G0 or the G1 and G2 renal-risk variants, and used Illumina human HT-12 v4 arrays and Affymetrix HTA 2.0 arrays to generate global gene expression data with doxycycline induction. Significantly altered pathways identified through bioinformatics analyses involved mitochondrial function; results from immunoblotting, immunofluorescence, and functional assays validated these findings. Overexpression of APOL1 by doxycycline induction in HEK293 Tet-on G1 and G2 cells led to impaired mitochondrial function, with markedly reduced maximum respiration rate, reserve respiration capacity, and mitochondrial membrane potential. Impaired mitochondrial function occurred before intracellular potassium depletion or reduced cell viability occurred. Analysis of global gene expression profiles in nondiseased primary proximal tubule cells from black patients revealed that the nicotinate phosphoribosyltransferase gene, responsible for NAD biosynthesis, was among the top downregulated transcripts in cells with two APOL1 renal-risk variants compared with those without renal-risk variants; nicotinate phosphoribosyltransferase also displayed gene expression patterns linked to mitochondrial dysfunction in HEK293 Tet-on APOL1 cell pathway analyses. These results suggest a pivotal role for mitochondrial dysfunction in APOL1-associated kidney disease.
Keywords: APOL1; chronic kidney disease; mitochondria.
Copyright © 2017 by the American Society of Nephrology.
Figures
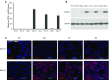
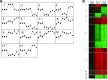
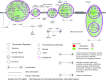

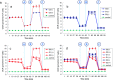


Comment in
-
Identifying the Intracellular Function of APOL1.J Am Soc Nephrol. 2017 Apr;28(4):1008-1011. doi: 10.1681/ASN.2016111262. Epub 2017 Feb 14. J Am Soc Nephrol. 2017. PMID: 28196842 Free PMC article. No abstract available.
References
-
- Genovese G, Friedman DJ, Ross MD, Lecordier L, Uzureau P, Freedman BI, Bowden DW, Langefeld CD, Oleksyk TK, Uscinski Knob AL, Bernhardy AJ, Hicks PJ, Nelson GW, Vanhollebeke B, Winkler CA, Kopp JB, Pays E, Pollak MR: Association of trypanolytic ApoL1 variants with kidney disease in African Americans. Science 329: 841–845, 2010 - PMC - PubMed
-
- Freedman BI, Langefeld CD, Andringa KK, Croker JA, Williams AH, Garner NE, Birmingham DJ, Hebert LA, Hicks PJ, Segal MS, Edberg JC, Brown EE, Alarcón GS, Costenbader KH, Comeau ME, Criswell LA, Harley JB, James JA, Kamen DL, Lim SS, Merrill JT, Sivils KL, Niewold TB, Patel NM, Petri M, Ramsey-Goldman R, Reveille JD, Salmon JE, Tsao BP, Gibson KL, Byers JR, Vinnikova AK, Lea JP, Julian BA, Kimberly RP; Lupus Nephritis–End‐Stage Renal Disease Consortium : End-stage renal disease in African Americans with lupus nephritis is associated with APOL1. Arthritis Rheumatol 66: 390–396, 2014 - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

