VAMP721 Conformations Unmask an Extended Motif for K+ Channel Binding and Gating Control
- PMID: 27821719
- PMCID: PMC5210753
- DOI: 10.1104/pp.16.01549
VAMP721 Conformations Unmask an Extended Motif for K+ Channel Binding and Gating Control
Abstract
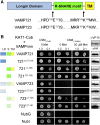
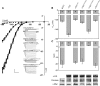
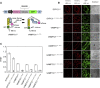
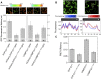
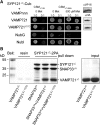
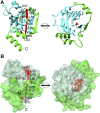
Soluble N-ethylmaleimide-sensitive factor attachment protein receptor (SNARE) proteins play a major role in membrane fusion and contribute to cell expansion, signaling, and polar growth in plants. The SNARE SYP121 of Arabidopsis thaliana that facilitates vesicle fusion at the plasma membrane also binds with, and regulates, K+ channels already present at the plasma membrane to affect K+ uptake and K+-dependent growth. Here, we report that its cognate partner VAMP721, which assembles with SYP121 to drive membrane fusion, binds to the KAT1 K+ channel via two sites on the protein, only one of which contributes to channel-gating control. Binding to the VAMP721 SNARE domain suppressed channel gating. By contrast, interaction with the amino-terminal longin domain conferred specificity on VAMP721 binding without influencing gating. Channel binding was defined by a linear motif within the longin domain. The SNARE domain is thought to wrap around this structure when not assembled with SYP121 in the SNARE complex. Fluorescence lifetime analysis showed that mutations within this motif, which suppressed channel binding and its effects on gating, also altered the conformational displacement between the VAMP721 SNARE and longin domains. The presence of these two channel-binding sites on VAMP721, one also required for SNARE complex assembly, implies a well-defined sequence of events coordinating K+ uptake and the final stages of vesicle traffic. It suggests that binding begins with VAMP721, and subsequently with SYP121, thereby coordinating K+ channel gating during SNARE assembly and vesicle fusion. Thus, our findings also are consistent with the idea that the K+ channels are nucleation points for SNARE complex assembly.
© 2017 American Society of Plant Biologists. All Rights Reserved.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
- BB/F001630/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/M01133X/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/M001601/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/I024496/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/L001276/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

