Engineering dynamical control of cell fate switching using synthetic phospho-regulons
- PMID: 27821768
- PMCID: PMC5127309
- DOI: 10.1073/pnas.1610973113
Engineering dynamical control of cell fate switching using synthetic phospho-regulons
Abstract
Many cells can sense and respond to time-varying stimuli, selectively triggering changes in cell fate only in response to inputs of a particular duration or frequency. A common motif in dynamically controlled cells is a dual-timescale regulatory network: although long-term fate decisions are ultimately controlled by a slow-timescale switch (e.g., gene expression), input signals are first processed by a fast-timescale signaling layer, which is hypothesized to filter what dynamic information is efficiently relayed downstream. Directly testing the design principles of how dual-timescale circuits control dynamic sensing, however, has been challenging, because most synthetic biology methods have focused solely on rewiring transcriptional circuits, which operate at a single slow timescale. Here, we report the development of a modular approach for flexibly engineering phosphorylation circuits using designed phospho-regulon motifs. By then linking rapid phospho-feedback with slower downstream transcription-based bistable switches, we can construct synthetic dual-timescale circuits in yeast in which the triggering dynamics and the end-state properties of the ON state can be selectively tuned. These phospho-regulon tools thus open up the possibility to engineer cells with customized dynamical control.
Keywords: dynamical control; phosphorylation; synthetic biology.
Conflict of interest statement
W.A.L. is a founder of Cell Design Labs and a member of its scientific advisory board.
Figures
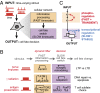



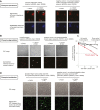


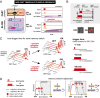
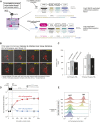



References
-
- Lucchesi W, Mizuno K, Giese KP. Novel insights into CaMKII function and regulation during memory formation. Brain Res Bull. 2011;85(1-2):2–8. - PubMed
-
- Kandel ER, Dudai Y, Mayford MR. The molecular and systems biology of memory. Cell. 2014;157(1):163–186. - PubMed
-
- Murphy LO, Smith S, Chen R-H, Fingar DC, Blenis J. Molecular interpretation of ERK signal duration by immediate early gene products. Nat Cell Biol. 2002;4(8):556–564. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

