Urban Transit System Microbial Communities Differ by Surface Type and Interaction with Humans and the Environment
- PMID: 27822528
- PMCID: PMC5069760
- DOI: 10.1128/mSystems.00018-16
Urban Transit System Microbial Communities Differ by Surface Type and Interaction with Humans and the Environment
Abstract
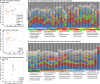
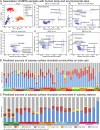
Public transit systems are ideal for studying the urban microbiome and interindividual community transfer. In this study, we used 16S amplicon and shotgun metagenomic sequencing to profile microbial communities on multiple transit surfaces across train lines and stations in the Boston metropolitan transit system. The greatest determinant of microbial community structure was the transit surface type. In contrast, little variation was observed between geographically distinct train lines and stations serving different demographics. All surfaces were dominated by human skin and oral commensals such as Propionibacterium, Corynebacterium, Staphylococcus, and Streptococcus. The detected taxa not associated with humans included generalists from alphaproteobacteria, which were especially abundant on outdoor touchscreens. Shotgun metagenomics further identified viral and eukaryotic microbes, including Propionibacterium phage and Malassezia globosa. Functional profiling showed that Propionibacterium acnes pathways such as propionate production and porphyrin synthesis were enriched on train holding surfaces (holds), while electron transport chain components for aerobic respiration were enriched on touchscreens and seats. Lastly, the transit environment was not found to be a reservoir of antimicrobial resistance and virulence genes. Our results suggest that microbial communities on transit surfaces are maintained from a metapopulation of human skin commensals and environmental generalists, with enrichments corresponding to local interactions with the human body and environmental exposures. IMPORTANCE Mass transit environments, specifically, urban subways, are distinct microbial environments with high occupant densities, diversities, and turnovers, and they are thus especially relevant to public health. Despite this, only three culture-independent subway studies have been performed, all since 2013 and all with widely differing designs and conclusions. In this study, we profiled the Boston subway system, which provides 238 million trips per year overseen by the Massachusetts Bay Transportation Authority (MBTA). This yielded the first high-precision microbial survey of a variety of surfaces, ridership environments, and microbiological functions (including tests for potential pathogenicity) in a mass transit environment. Characterizing microbial profiles for multiple transit systems will become increasingly important for biosurveillance of antibiotic resistance genes or pathogens, which can be early indicators for outbreak or sanitation events. Understanding how human contact, materials, and the environment affect microbial profiles may eventually allow us to rationally design public spaces to sustain our health in the presence of microbial reservoirs. Author Video: An author video summary of this article is available.
Keywords: built environment; microbiome; subway; transit.
Figures





Comment in
-
Avoiding Pandemic Fears in the Subway and Conquering the Platypus.mSystems. 2016 Jun 28;1(3):e00050-16. doi: 10.1128/mSystems.00050-16. eCollection 2016 May-Jun. mSystems. 2016. PMID: 27832215 Free PMC article.
References
-
- National Transit Database 2015. Monthly module raw data release. http://www.ntdprogram.gov/ntdprogram/data.htm.
-
- Lax S, Smith DP, Hampton-Marcell J, Owens SM, Handley KM, Scott NM, Gibbons SM, Larsen P, Shogan BD, Weiss S, Metcalf JL, Ursell LK, Vázquez-Baeza Y, Van Treuren W, Hasan NA, Gibson MK, Colwell R, Dantas G, Knight R, Gilbert JA. 2014. Longitudinal analysis of microbial interaction between humans and the indoor environment. Science 345:1048–1052. doi: 10.1126/science.1254529. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
