The Oral and Skin Microbiomes of Captive Komodo Dragons Are Significantly Shared with Their Habitat
- PMID: 27822543
- PMCID: PMC5069958
- DOI: 10.1128/mSystems.00046-16
The Oral and Skin Microbiomes of Captive Komodo Dragons Are Significantly Shared with Their Habitat
Abstract
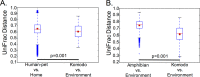
Examining the way in which animals, including those in captivity, interact with their environment is extremely important for studying ecological processes and developing sophisticated animal husbandry. Here we use the Komodo dragon (Varanus komodoensis) to quantify the degree of sharing of salivary, skin, and fecal microbiota with their environment in captivity. Both species richness and microbial community composition of most surfaces in the Komodo dragon's environment are similar to the Komodo dragon's salivary and skin microbiota but less similar to the stool-associated microbiota. We additionally compared host-environment microbiome sharing between captive Komodo dragons and their enclosures, humans and pets and their homes, and wild amphibians and their environments. We observed similar host-environment microbiome sharing patterns among humans and their pets and Komodo dragons, with high levels of human/pet- and Komodo dragon-associated microbes on home and enclosure surfaces. In contrast, only small amounts of amphibian-associated microbes were detected in the animals' environments. We suggest that the degree of sharing between the Komodo dragon microbiota and its enclosure surfaces has important implications for animal health. These animals evolved in the context of constant exposure to a complex environmental microbiota, which likely shaped their physiological development; in captivity, these animals will not receive significant exposure to microbes not already in their enclosure, with unknown consequences for their health. IMPORTANCE Animals, including humans, have evolved in the context of exposure to a variety of microbial organisms present in the environment. Only recently have humans, and some animals, begun to spend a significant amount of time in enclosed artificial environments, rather than in the more natural spaces in which most of evolution took place. The consequences of this radical change in lifestyle likely extend to the microbes residing in and on our bodies and may have important implications for health and disease. A full characterization of host-microbe sharing in both closed and open environments will provide crucial information that may enable the improvement of health in humans and in captive animals, both of which experience a greater incidence of disease (including chronic illness) than counterparts living under more ecologically natural conditions.
Keywords: Komodo dragon; SourceTracker; built environment; human microbiome; microbiome.
Figures



References
-
- McFall-Ngai M, Hadfield MG, Bosch TCG, Carey HV, Domazet-Lošo T, Douglas AE, Dubilier N, Eberl G, Fukami T, Gilbert SF, Hentschel U, King N, Kjelleberg S, Knoll AH, Kremer N, Mazmanian SK, Metcalf JL, Nealson K, Pierce NE, Rawls JF, Reid A, Ruby EG, Rumpho M, Sanders JG, Tautz D, Wernegreen JJ. 2013. Animals in a bacterial world, a new imperative for the life sciences. Proc Natl Acad Sci U S A 110:3229–3236. doi: 10.1073/pnas.1218525110. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
