Genomic analyses identify recurrent MEF2D fusions in acute lymphoblastic leukaemia
- PMID: 27824051
- PMCID: PMC5105166
- DOI: 10.1038/ncomms13331
Genomic analyses identify recurrent MEF2D fusions in acute lymphoblastic leukaemia
Abstract
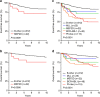
Chromosomal rearrangements are initiating events in acute lymphoblastic leukaemia (ALL). Here using RNA sequencing of 560 ALL cases, we identify rearrangements between MEF2D (myocyte enhancer factor 2D) and five genes (BCL9, CSF1R, DAZAP1, HNRNPUL1 and SS18) in 22 B progenitor ALL (B-ALL) cases with a distinct gene expression profile, the most common of which is MEF2D-BCL9. Examination of an extended cohort of 1,164 B-ALL cases identified 30 cases with MEF2D rearrangements, which include an additional fusion partner, FOXJ2; thus, MEF2D-rearranged cases comprise 5.3% of cases lacking recurring alterations. MEF2D-rearranged ALL is characterized by a distinct immunophenotype, DNA copy number alterations at the rearrangement sites, older diagnosis age and poor outcome. The rearrangements result in enhanced MEF2D transcriptional activity, lymphoid transformation, activation of HDAC9 expression and sensitive to histone deacetylase inhibitor treatment. Thus, MEF2D-rearranged ALL represents a distinct form of high-risk leukaemia, for which new therapeutic approaches should be considered.
Figures



References
-
- Hunger S. P. & Mullighan C. G. Acute lymphoblastic leukemia in children. N. Engl. J. Med. 373, 1541–1552 (2015). - PubMed
-
- Moorman A. V. The clinical relevance of chromosomal and genomic abnormalities in B-cell precursor acute lymphoblastic leukaemia. Blood Rev. 26, 123–135 (2012). - PubMed
-
- Willis T. G. et al. Molecular cloning of translocation t(1;14)(q21;q32) defines a novel gene (BCL9) at chromosome 1q21. Blood 91, 1873–1881 (1998). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- U10 CA180861/CA/NCI NIH HHS/United States
- 21019/CRUK_/Cancer Research UK/United Kingdom
- U24 CA196172/CA/NCI NIH HHS/United States
- U10 CA180850/CA/NCI NIH HHS/United States
- P30 CA021765/CA/NCI NIH HHS/United States
- U01 CA157937/CA/NCI NIH HHS/United States
- U10 CA180899/CA/NCI NIH HHS/United States
- HHSN261200800001C/RC/CCR NIH HHS/United States
- U24 CA114737/CA/NCI NIH HHS/United States
- HHSN261200800001E/CA/NCI NIH HHS/United States
- U10 CA098413/CA/NCI NIH HHS/United States
- U24 CA114766/CA/NCI NIH HHS/United States
- U24 CA196171/CA/NCI NIH HHS/United States
- U10 CA098543/CA/NCI NIH HHS/United States
- RC1 CA145707/CA/NCI NIH HHS/United States
- U10 CA180820/CA/NCI NIH HHS/United States
- U10 CA180827/CA/NCI NIH HHS/United States
- P50 GM115279/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

