De novo genic mutations among a Chinese autism spectrum disorder cohort
- PMID: 27824329
- PMCID: PMC5105161
- DOI: 10.1038/ncomms13316
De novo genic mutations among a Chinese autism spectrum disorder cohort
Abstract
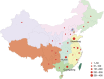
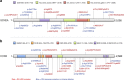
Recurrent de novo (DN) and likely gene-disruptive (LGD) mutations contribute significantly to autism spectrum disorders (ASDs) but have been primarily investigated in European cohorts. Here, we sequence 189 risk genes in 1,543 Chinese ASD probands (1,045 from trios). We report an 11-fold increase in the odds of DN LGD mutations compared with expectation under an exome-wide neutral model of mutation. In aggregate, ∼4% of ASD patients carry a DN mutation in one of just 29 autism risk genes. The most prevalent gene for recurrent DN mutations is SCN2A (1.1% of patients) followed by CHD8, DSCAM, MECP2, POGZ, WDFY3 and ASH1L. We identify novel DN LGD recurrences (GIGYF2, MYT1L, CUL3, DOCK8 and ZNF292) and DN mutations in previous ASD candidates (ARHGAP32, NCOR1, PHIP, STXBP1, CDKL5 and SHANK1). Phenotypic follow-up confirms potential subtypes and highlights how large global cohorts might be leveraged to prove the pathogenic significance of individually rare mutations.
Conflict of interest statement
E.E.E. is on the scientific advisory board (SAB) of DNAnexus, Inc. and was an SAB member of Pacific Biosciences, Inc. (2009–2013) and SynapDx Corp. (2011–2013); E.E.E. is a consultant for Kunming University of Science and Technology (KUST) as part of the 1,000 China Talent Program. The other authors declare no competing financial interests.
Figures
References
-
- Lai M. C., Lombardo M. V. & Baron-Cohen S. Autism. Lancet 383, 896–910 (2014). - PubMed
-
- Developmental Disabilities Monitoring Network Surveillance Year Principal I, Centers for Disease C, Prevention. Prevalence of autism spectrum disorder among children aged 8 years - autism and developmental disabilities monitoring network, 11 sites, United States, 2010. MMWR Surveill. Summ. 63, 1–21 (2014). - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

