Critical decisions in metaproteomics: achieving high confidence protein annotations in a sea of unknowns
- PMID: 27824341
- PMCID: PMC5270573
- DOI: 10.1038/ismej.2016.132
Critical decisions in metaproteomics: achieving high confidence protein annotations in a sea of unknowns
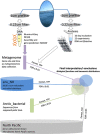
Figures


References
-
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. (1990). Basic local alignment search tool. J Mol Biol 215: 403–410. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

