Membrane vesicles in sea water: heterogeneous DNA content and implications for viral abundance estimates
- PMID: 27824343
- PMCID: PMC5270575
- DOI: 10.1038/ismej.2016.134
Membrane vesicles in sea water: heterogeneous DNA content and implications for viral abundance estimates
Abstract
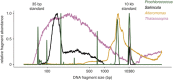
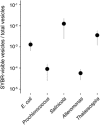
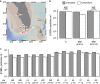
Diverse microbes release membrane-bound extracellular vesicles from their outer surfaces into the surrounding environment. Vesicles are found in numerous habitats including the oceans, where they likely have a variety of functional roles in microbial ecosystems. Extracellular vesicles are known to contain a range of biomolecules including DNA, but the frequency with which DNA is packaged in vesicles is unknown. Here, we examine the quantity and distribution of DNA associated with vesicles released from five different bacteria. The average quantity of double-stranded DNA and size distribution of DNA fragments released within vesicles varies among different taxa. Although some vesicles contain sufficient DNA to be visible following staining with the SYBR fluorescent DNA dyes typically used to enumerate viruses, this represents only a small proportion (<0.01-1%) of vesicles. Thus DNA is packaged heterogeneously within vesicle populations, and it appears that vesicles are likely to be a minor component of SYBR-visible particles in natural sea water compared with viruses. Consistent with this hypothesis, chloroform treatment of coastal and offshore seawater samples reveals that vesicles increase epifluorescence-based particle (viral) counts by less than an order of magnitude and their impact is variable in space and time.
Figures
References
-
- Ackermann H-W, DuBow MS. (1987) Viruses of Prokaryotes. CRC Press: Boca Raton, FL, USA.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

