Sequential and counter-selectable cassettes for fission yeast
- PMID: 27825338
- PMCID: PMC5101803
- DOI: 10.1186/s12896-016-0307-4
Sequential and counter-selectable cassettes for fission yeast
Abstract
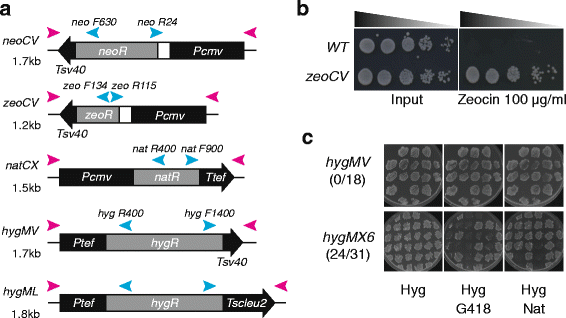
Background: Fission yeast is one of the most commonly used model organisms for studying genetics. For selection of desirable genotypes, antibiotic resistance cassettes are widely integrated into the genome near genes of interest. In yeasts, this is achieved by PCR amplification of the cassette flanked by short homology sequences, which can be incorporated by homology directed repair. However, the currently available cassettes all share the same tef promoter and terminator sequences. It can therefore be challenging to perform multiple genetic modifications by PCR-based targeting, as existing resistance cassettes in strains can be favored for recombination due to shared homology between the cassettes.
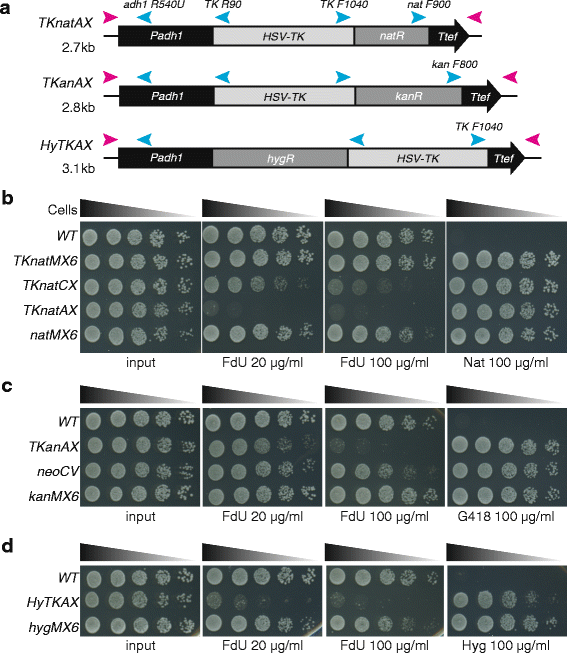
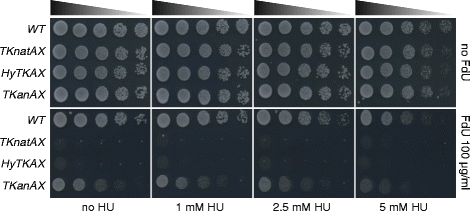
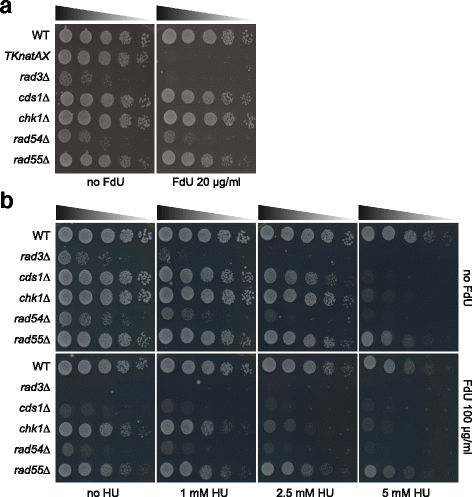
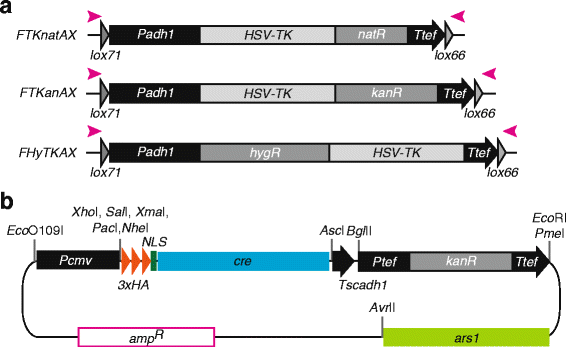
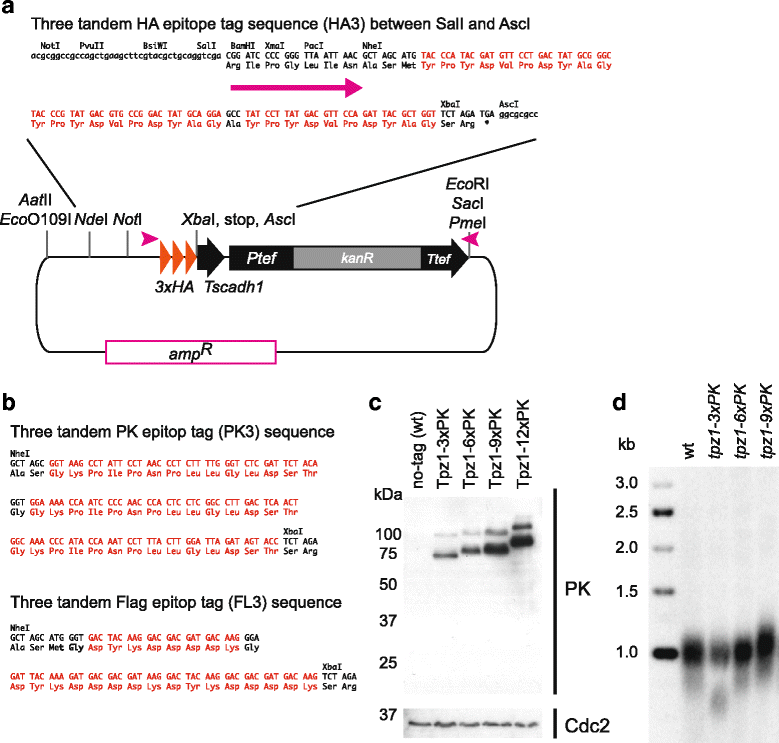
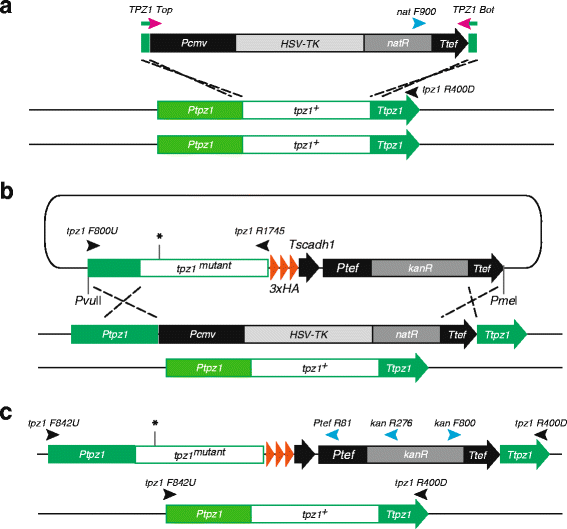
Results: Here we have generated new selection cassettes that do not recombine with those traditionally used. We achieved this by swapping the tef promoter and terminator sequences in the established antibiotic resistance MX6 cassette series for alternative promoters and/or terminators. The newly created selection cassettes did not recombine with the tef-containing MX6 cassettes already present in the genome, allowing for sequential gene targeting using the PCR-based method. In addition, we have generated a series of plasmids to facilitate the C-terminal tagging of genes with desired epitopes. We also utilized the anti-selection gene HSV-TK, which results in cell death in strains grown on the drug 5-Fluoro-2'-deoxyuridine (FdU, Floxuridin or FUDR). By fusing an antibiotic resistance gene to HSV-TK, we were able to select on the relevant antibiotic as well as counter-select on FdU media to confirm the desired genomic modification had been made. We noted that the efficiency of the counter selection by FdU was enhanced by treatment with hydroxyurea. However, a number of DNA replication checkpoint and homologous recombination mutants, including rad3∆, cds1∆, rad54∆ and rad55∆, exhibited sensitivity to FdU even though those strains did not carry the HSV-TK gene. To remove counter-selectable markers, we introduced the Cre-loxP irreversible recombination method. Finally, utilizing the negative selectable markers, we showed efficient induction of point mutations in an endogenous gene by a two-step transformation method.
Conclusions: The plasmid constructs and techniques described here are invaluable tools for sequential gene targeting and will simplify construction of fission yeast strains required for study.
Keywords: DNA replication; FUdR; Gene disruption and insertion; HA, Flag, PK tagging; Point mutation; Schizosaccharomyces pombe; Thymidine kinase; Zeocin.
Figures
References
-
- Bahler J, Wu JQ, Longtine MS, Shah NG, McKenzie A, 3rd, Steever AB, Wach A, Philippsen P, Pringle JR. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast. 1998;14(10):943–951. doi: 10.1002/(SICI)1097-0061(199807)14:10<943::AID-YEA292>3.0.CO;2-Y. - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

