DNA Base Flipping: A General Mechanism for Writing, Reading, and Erasing DNA Modifications
- PMID: 27826845
- PMCID: PMC5542066
- DOI: 10.1007/978-3-319-43624-1_14
DNA Base Flipping: A General Mechanism for Writing, Reading, and Erasing DNA Modifications
Abstract
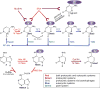
The modification of DNA bases is a classic hallmark of epigenetics. Four forms of modified cytosine-5-methylcytosine, 5-hydroxymethylcytosine, 5-formylcytosine, and 5-carboxylcytosine-have been discovered in eukaryotic DNA. In addition to cytosine carbon-5 modifications, cytosine and adenine methylated in the exocyclic amine-N4-methylcytosine and N6-methyladenine-are other modified DNA bases discovered even earlier. Each modified base can be considered a distinct epigenetic signal with broader biological implications beyond simple chemical changes. Since 1994, crystal structures of proteins and enzymes involved in writing, reading, and erasing modified bases have become available. Here, we present a structural synopsis of writers, readers, and erasers of the modified bases from prokaryotes and eukaryotes. Despite significant differences in structures and functions, they are remarkably similar regarding their engagement in flipping a target base/nucleotide within DNA for specific recognitions and/or reactions. We thus highlight base flipping as a common structural framework broadly applied by distinct classes of proteins and enzymes across phyla for epigenetic regulations of DNA.
Figures

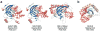
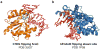
References
-
- Arita K, Ariyoshi M, Tochio H, Nakamura Y, Shirakawa M. Recognition of hemi-methylated DNA by the SRA protein UHRF1 by a base-flipping mechanism. Nature. 2008;455(7214):818–21. - PubMed
-
- Avvakumov GV, Walker JR, Xue S, Li Y, Duan S, Bronner C, et al. Structural basis for recognition of hemi-methylated DNA by the SRA domain of human UHRF1. Nature. 2008;455(7214):822–5. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

