Impact of mutational profiles on response of primary oestrogen receptor-positive breast cancers to oestrogen deprivation
- PMID: 27827358
- PMCID: PMC5105193
- DOI: 10.1038/ncomms13294
Impact of mutational profiles on response of primary oestrogen receptor-positive breast cancers to oestrogen deprivation
Abstract
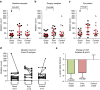
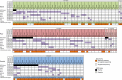
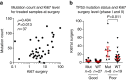
Pre-surgical studies allow study of the relationship between mutations and response of oestrogen receptor-positive (ER+) breast cancer to aromatase inhibitors (AIs) but have been limited to small biopsies. Here in phase I of this study, we perform exome sequencing on baseline, surgical core-cuts and blood from 60 patients (40 AI treated, 20 controls). In poor responders (based on Ki67 change), we find significantly more somatic mutations than good responders. Subclones exclusive to baseline or surgical cores occur in ∼30% of tumours. In phase II, we combine targeted sequencing on another 28 treated patients with phase I. We find six genes frequently mutated: PIK3CA, TP53, CDH1, MLL3, ABCA13 and FLG with 71% concordance between paired cores. TP53 mutations are associated with poor response. We conclude that multiple biopsies are essential for confident mutational profiling of ER+ breast cancer and TP53 mutations are associated with resistance to oestrogen deprivation therapy.
Conflict of interest statement
J.R. is employed by Oncimmune. C.H. is a paid consultant to Genomic Health, M.D. is a paid consultant to Pfizer and NanoString. J.R. holds stock in Oncimmune and FaHRAS. C.H. received travel expenses from Novartis, and J.R. from Oncimmune, AstraZeneca, Bayer, Novartis and Syndax. J.R. received honoraria from AstraZeneca, Bayer and Amgen. J.R. receives research funding from Oncimmune, and M.D. from Novartis, AstraZeneca and Pfizer. J.R. holds patents from Oncimmune and is in the speakers' bureau of AstraZeneca. The remaining authors declare no competing financial interests.
Figures


References
-
- Dowsett M. et al.. Endocrine therapy, new biologicals, and new study designs for presurgical studies in breast cancer. J. Natl. Cancer. Inst. Monogr. 2011, 120–123 (2011). - PubMed
-
- Cancer.Net. Breast cancer - overview. Available at http://www.cancer.net/cancer-types/breast-cancer/overview (2016).
-
- Early Breast Cancer Trialists' Collaborative Group. Aromatase inhibitors versus tamoxifen in early breast cancer: patient-level meta-analysis of the randomised trials. Lancet 386, 1341–1352 (2015). - PubMed
-
- Smith I. E. & Dowsett M. Aromatase inhibitors in breast cancer. N. Engl. J. Med. 348, 2431–2442 (2003). - PubMed
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

