FANCD2 functions as a critical factor downstream of MiTF to maintain the proliferation and survival of melanoma cells
- PMID: 27827420
- PMCID: PMC5101529
- DOI: 10.1038/srep36539
FANCD2 functions as a critical factor downstream of MiTF to maintain the proliferation and survival of melanoma cells
Abstract
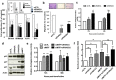
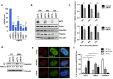
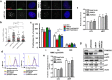
Proteins involved in genetic stability maintenance and safeguarding DNA replication act not only against cancer initiation but could also play a major role in sustaining cancer progression. Here, we report that the FANC pathway is highly expressed in metastatic melanoma harboring the oncogenic microphthalmia-associated transcription factor (MiTF). We show that MiTF downregulation in melanoma cells lowers the expression of several FANC genes and proteins. Moreover, we observe that, similarly to the consequence of MiTF downregulation, FANC pathway silencing alters proliferation, migration and senescence of human melanoma cells. We demonstrate that the FANC pathway acts downstream MiTF and establish the existence of an epistatic relationship between MiTF and the FANC pathway. Our findings point to a central role of the FANC pathway in cellular and chromosomal resistance to both DNA damage and targeted therapies in melanoma cells. Thus, the FANC pathway is a promising new therapeutic target in melanoma treatment.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

