Proteomic Landscape of Tissue-Specific Cyclin E Functions in Vivo
- PMID: 27828963
- PMCID: PMC5102403
- DOI: 10.1371/journal.pgen.1006429
Proteomic Landscape of Tissue-Specific Cyclin E Functions in Vivo
Abstract
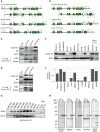
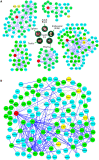
E-type cyclins (cyclins E1 and E2) are components of the cell cycle machinery that has been conserved from yeast to humans. The major function of E-type cyclins is to drive cell division. It is unknown whether in addition to their 'core' cell cycle functions, E-type cyclins also perform unique tissue-specific roles. Here, we applied high-throughput mass spectrometric analyses of mouse organs to define the repertoire of cyclin E protein partners in vivo. We found that cyclin E interacts with distinct sets of proteins in different compartments. These cyclin E interactors are highly enriched for phosphorylation targets of cyclin E and its catalytic partner, the cyclin-dependent kinase 2 (Cdk2). Among cyclin E interactors we identified several novel tissue-specific substrates of cyclin E-Cdk2 kinase. In proliferating compartments, cyclin E-Cdk2 phosphorylates Lin proteins within the DREAM complex. In the testes, cyclin E-Cdk2 phosphorylates Mybl1 and Dmrtc2, two meiotic transcription factors that represent key regulators of spermatogenesis. In embryonic and adult brains cyclin E interacts with proteins involved in neurogenesis, while in adult brains also with proteins regulating microtubule-based processes and microtubule cytoskeleton. We also used quantitative proteomics to demonstrate re-wiring of the cyclin E interactome upon ablation of Cdk2. This approach can be used to study how protein interactome changes during development or in any pathological state such as aging or cancer.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures





Similar articles
-
Kinase-independent function of E-type cyclins in liver cancer.Proc Natl Acad Sci U S A. 2018 Jan 30;115(5):1015-1020. doi: 10.1073/pnas.1711477115. Epub 2018 Jan 16. Proc Natl Acad Sci U S A. 2018. PMID: 29339491 Free PMC article.
-
Mammalian E-type cyclins control chromosome pairing, telomere stability and CDK2 localization in male meiosis.PLoS Genet. 2014 Feb 27;10(2):e1004165. doi: 10.1371/journal.pgen.1004165. eCollection 2014 Feb. PLoS Genet. 2014. PMID: 24586195 Free PMC article.
-
Concurrent deletion of cyclin E1 and cyclin-dependent kinase 2 in hepatocytes inhibits DNA replication and liver regeneration in mice.Hepatology. 2014 Feb;59(2):651-60. doi: 10.1002/hep.26584. Epub 2013 Dec 24. Hepatology. 2014. PMID: 23787781 Free PMC article.
-
Cyclin E in normal physiology and disease states.Trends Cell Biol. 2021 Sep;31(9):732-746. doi: 10.1016/j.tcb.2021.05.001. Epub 2021 May 27. Trends Cell Biol. 2021. PMID: 34052101 Free PMC article. Review.
-
Cyclins and CDKS in development and cancer: lessons from genetically modified mice.Front Biosci. 2006 Jan 1;11:1164-88. doi: 10.2741/1871. Front Biosci. 2006. PMID: 16146805 Review.
Cited by
-
Meiotic gatekeeper STRA8 regulates cell cycle by interacting with SETD8 during spermatogenesis.J Cell Mol Med. 2020 Apr;24(7):4194-4211. doi: 10.1111/jcmm.15080. Epub 2020 Feb 24. J Cell Mol Med. 2020. PMID: 32090428 Free PMC article.
-
MYC Modulation around the CDK2/p27/SKP2 Axis.Genes (Basel). 2017 Jun 30;8(7):174. doi: 10.3390/genes8070174. Genes (Basel). 2017. PMID: 28665315 Free PMC article. Review.
-
Functional Analysis of the Cyclin E Gene in the Reproductive Development of Rainbow Trout (Oncorhynchus mykiss).Biology (Basel). 2025 Jul 16;14(7):862. doi: 10.3390/biology14070862. Biology (Basel). 2025. PMID: 40723419 Free PMC article.
-
Tandem immuno-purification of affinity-tagged proteins from mouse testis extracts for MS analysis.STAR Protoc. 2022 Jun 13;3(2):101452. doi: 10.1016/j.xpro.2022.101452. eCollection 2022 Jun 17. STAR Protoc. 2022. PMID: 35719267 Free PMC article.
-
Emerging Role of the DREAM Complex in Cancer and Therapeutic Opportunities.Int J Mol Sci. 2025 Jan 1;26(1):322. doi: 10.3390/ijms26010322. Int J Mol Sci. 2025. PMID: 39796178 Free PMC article. Review.
References
-
- Geng Y, Yu Q, Sicinska E, Das M, Schneider JE, et al. (2003) Cyclin E ablation in the mouse. Cell 114: 431–443. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

