Remarkable Diversity and Prevalence of Dagger Nematodes of the Genus Xiphinema Cobb, 1913 (Nematoda: Longidoridae) in Olives Revealed by Integrative Approaches
- PMID: 27829048
- PMCID: PMC5102458
- DOI: 10.1371/journal.pone.0165412
Remarkable Diversity and Prevalence of Dagger Nematodes of the Genus Xiphinema Cobb, 1913 (Nematoda: Longidoridae) in Olives Revealed by Integrative Approaches
Abstract
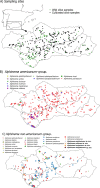
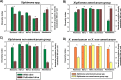
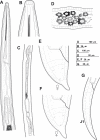
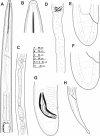
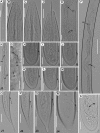
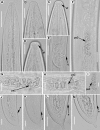
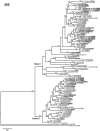
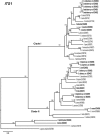
The genus Xiphinema includes a remarkable group of invertebrates of the phylum Nematoda comprising ectoparasitic animals of many wild and cultivated plants. Damage is caused by direct feeding on root cells and by vectoring nepoviruses that cause diseases on several crops. Precise identification of Xiphinema species is critical for launching appropriate control measures. We make available the first detailed information on the diversity and distribution of Xiphinema species infesting wild and cultivated olive in a wide-region in southern Spain that included 211 locations from which 453 sampling sites were analyzed. The present study identified thirty-two Xiphinema spp. in the rhizosphere of olive trees, ten species belonging to Xiphinema americanum-group, whereas twenty-two were attributed to Xiphinema non-americanum-group. These results increase our current knowledge on the biodiversity of Xiphinema species identified in olives and include the description of four new species (Xiphinema andalusiense sp. nov., Xiphinema celtiense sp. nov., Xiphinema iznajarense sp. nov., and Xiphinema mengibarense sp. nov.), and two new records for cultivate olives (X. cadavalense and X. conurum). We also found evidence of remarkable prevalence of Xiphinema spp. in olive trees, viz. 85.0% (385 out of 453 sampling sites), and they were widely distributed in both wild and cultivated olives, with 26 and 17 Xiphinema spp., respectively. Diversity indexes (Richness, Hill´s diversity, Hill´s reciprocal of D and Hill´s evenness) were significantly affected by olive type. We also developed a comparative morphological and morphometrical study together with molecular data from three nuclear ribosomal RNA genes (D2-D3 expansion segments of 28S, ITS1, and partial 18S). Molecular characterization and phylogenetic analyses allowed the delimitation and discrimination of four new species of the genus described herein and three known species. Phylogenetic analyses of Xiphinema spp. resulted in a general consensus of these species groups. This study is the most complete phylogenetic analysis for Xiphinema non-americanum-group species to date.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures









References
-
- Wolters V. Biodiversity of soil animals and its function. European Journal of Soil Biology. 2001;37:221–7.
-
- Decaens T, Jiménez JJ, Gioia C, Measey GJ, Lavelle P. The values of soil animals for conservation biology. European Journal of Soil Biology. 2006;42:23–8.
-
- Bongers T, Ferris H. Nematode community structure as a bioindicator in environmental monitoring. Trends in Ecology and Evolution. 1999;14:224–8. - PubMed
-
- Wyss U. Root parasitic nematodes: an overview In: FMWG C. Fenoll, Ohl S.A., editor. Cellular and molecularaspects of plant-nematode interactions. 10. Dordrecht, The Netherlands: Kluwer Academic Publisher; 1997. p. 5–22.
-
- Decraemer W, Hunt D. Structure and clasiffication In: Perry RN, Moens M., editor. Plant Nematology. Wallingford, UK: CABI; 2006. p. 3–32.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

