Theranostic barcoded nanoparticles for personalized cancer medicine
- PMID: 27830705
- PMCID: PMC5109543
- DOI: 10.1038/ncomms13325
Theranostic barcoded nanoparticles for personalized cancer medicine
Abstract
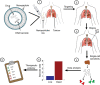
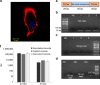
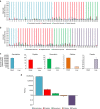
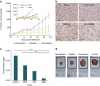
Personalized medicine promises to revolutionize cancer therapy by matching the most effective treatment to the individual patient. Using a nanoparticle-based system, we predict the therapeutic potency of anticancer medicines in a personalized manner. We carry out the diagnostic stage through a multidrug screen performed inside the tumour, extracting drug activity information with single cell sensitivity. By using 100 nm liposomes, loaded with various cancer drugs and corresponding synthetic DNA barcodes, we find a correlation between the cell viability and the drug it was exposed to, according to the matching barcodes. Based on this screen, we devise a treatment protocol for mice bearing triple-negative breast-cancer tumours, and its results confirm the diagnostic prediction. We show that the use of nanotechnology in cancer care is effective for generating personalized treatment protocols.
Figures

Comment in
-
Taking cancer drug screening very personally.Sci Transl Med. 2016 Nov 30;8(367):367ec190. doi: 10.1126/scitranslmed.aal2799. Sci Transl Med. 2016. PMID: 27903858 No abstract available.
References
-
- Symmans W. F. et al.. Measurement of residual breast cancer burden to predict survival after neoadjuvant chemotherapy. J. Clin. Oncol. 25, 4414–4422 (2007). - PubMed
-
- Thomas E. et al.. The use of alternate, non-cross-resistant adjuvant chemotherapy on the basis of pathologic response to a neoadjuvant doxorubicin-based regimen in women with operable breast cancer: long-term results from a prospective randomized trial. J. Clin. Oncol. 22, 2294–2302 (2004). - PubMed
-
- Buzdar A. U. et al.. Prospective evaluation of paclitaxel versus combination chemotherapy with fluorouracil, doxorubicin, and cyclophosphamide as neoadjuvant therapy in patients with operable breast cancer. J. Clin. Oncol. 17, 3412–3417 (1999). - PubMed
-
- Sotiriou C. & Piccart M. J. Taking gene-expression profiling to the clinic: when will molecular signatures become relevant to patient care? Nat. Rev. Cancer. 7, 545–553 (2007). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

