Evolution of Osteocrin as an activity-regulated factor in the primate brain
- PMID: 27830782
- PMCID: PMC5499253
- DOI: 10.1038/nature20111
Evolution of Osteocrin as an activity-regulated factor in the primate brain
Abstract
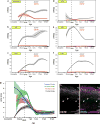
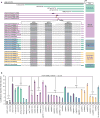
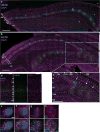
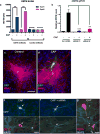
Sensory stimuli drive the maturation and function of the mammalian nervous system in part through the activation of gene expression networks that regulate synapse development and plasticity. These networks have primarily been studied in mice, and it is not known whether there are species- or clade-specific activity-regulated genes that control features of brain development and function. Here we use transcriptional profiling of human fetal brain cultures to identify an activity-dependent secreted factor, Osteocrin (OSTN), that is induced by membrane depolarization of human but not mouse neurons. We find that OSTN has been repurposed in primates through the evolutionary acquisition of DNA regulatory elements that bind the activity-regulated transcription factor MEF2. In addition, we demonstrate that OSTN is expressed in primate neocortex and restricts activity-dependent dendritic growth in human neurons. These findings suggest that, in response to sensory input, OSTN regulates features of neuronal structure and function that are unique to primates.
Conflict of interest statement
The authors declare no competing financial interests. Readers are welcome to comment on the online version of the paper.
Figures
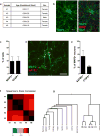
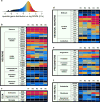
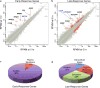

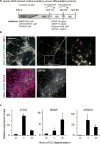

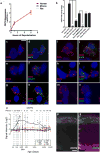

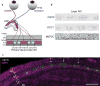

Comment in
-
Evolution: Genomic remodelling in the primate brain.Nature. 2016 Nov 10;539(7628):171-172. doi: 10.1038/539171a. Nature. 2016. PMID: 27830798 No abstract available.
References
-
- Bufill E, Agustí J, Blesa R. Human neoteny revisited: The case of synaptic plasticity. Am J Hum Biol. 2011;23:729–739. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

