Comparative Bioinformatics Analysis of Transcription Factor Genes Indicates Conservation of Key Regulatory Domains among Babesia bovis, Babesia microti, and Theileria equi
- PMID: 27832060
- PMCID: PMC5104403
- DOI: 10.1371/journal.pntd.0004983
Comparative Bioinformatics Analysis of Transcription Factor Genes Indicates Conservation of Key Regulatory Domains among Babesia bovis, Babesia microti, and Theileria equi
Abstract
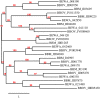
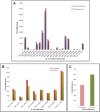
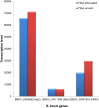
Apicomplexa tick-borne hemoparasites, including Babesia bovis, Babesia microti, and Theileria equi are responsible for bovine and human babesiosis and equine theileriosis, respectively. These parasites of vast medical, epidemiological, and economic impact have complex life cycles in their vertebrate and tick hosts. Large gaps in knowledge concerning the mechanisms used by these parasites for gene regulation remain. Regulatory genes coding for DNA binding proteins such as members of the Api-AP2, HMG, and Myb families are known to play crucial roles as transcription factors. Although the repertoire of Api-AP2 has been defined and a HMG gene was previously identified in the B. bovis genome, these regulatory genes have not been described in detail in B. microti and T. equi. In this study, comparative bioinformatics was used to: (i) identify and map genes encoding for these transcription factors among three parasites' genomes; (ii) identify a previously unreported HMG gene in B. microti; (iii) define a repertoire of eight conserved Myb genes; and (iv) identify AP2 correlates among B. bovis and the better-studied Plasmodium parasites. Searching the available transcriptome of B. bovis defined patterns of transcription of these three gene families in B. bovis erythrocyte stage parasites. Sequence comparisons show conservation of functional domains and general architecture in the AP2, Myb, and HMG proteins, which may be significant for the regulation of common critical parasite life cycle transitions in B. bovis, B. microti, and T. equi. A detailed understanding of the role of gene families encoding DNA binding proteins will provide new tools for unraveling regulatory mechanisms involved in B. bovis, B. microti, and T. equi life cycles and environmental adaptive responses and potentially contributes to the development of novel convergent strategies for improved control of babesiosis and equine piroplasmosis.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures





Similar articles
-
Assessment of Draxxin® (tulathromycin) as an inhibitor of in vitro growth of Babesia bovis, Babesia bigemina and Theileria equi.Int J Parasitol Drugs Drug Resist. 2018 Aug;8(2):265-270. doi: 10.1016/j.ijpddr.2018.04.004. Epub 2018 Apr 17. Int J Parasitol Drugs Drug Resist. 2018. PMID: 29689532 Free PMC article.
-
Differential expression of three members of the multidomain adhesion CCp family in Babesia bigemina, Babesia bovis and Theileria equi.PLoS One. 2013 Jul 3;8(7):e67765. doi: 10.1371/journal.pone.0067765. Print 2013. PLoS One. 2013. PMID: 23844089 Free PMC article.
-
Endochin-like quinolone-300 and ELQ-316 inhibit Babesia bovis, B. bigemina, B. caballi and Theileria equi.Parasit Vectors. 2020 Dec 3;13(1):606. doi: 10.1186/s13071-020-04487-3. Parasit Vectors. 2020. PMID: 33272316 Free PMC article.
-
Review of equine piroplasmosis.J Vet Intern Med. 2013 Nov-Dec;27(6):1334-46. doi: 10.1111/jvim.12168. Epub 2013 Aug 28. J Vet Intern Med. 2013. PMID: 24033559 Review.
-
Vector ecology of equine piroplasmosis.Annu Rev Entomol. 2015 Jan 7;60:561-80. doi: 10.1146/annurev-ento-010814-021110. Annu Rev Entomol. 2015. PMID: 25564746 Review.
Cited by
-
Thrombospondin-Related Anonymous Protein (TRAP) Family Expression by Babesia bovis Life Stages within the Mammalian Host and Tick Vector.Microorganisms. 2022 Nov 2;10(11):2173. doi: 10.3390/microorganisms10112173. Microorganisms. 2022. PMID: 36363765 Free PMC article.
-
Vaccination of cattle with the Babesia bovis sexual-stage protein HAP2 abrogates parasite transmission by Rhipicephalus microplus ticks.NPJ Vaccines. 2023 Sep 27;8(1):140. doi: 10.1038/s41541-023-00741-8. NPJ Vaccines. 2023. PMID: 37758790 Free PMC article.
-
Unravelling the cellular and molecular pathogenesis of bovine babesiosis: is the sky the limit?Int J Parasitol. 2019 Feb;49(2):183-197. doi: 10.1016/j.ijpara.2018.11.002. Epub 2019 Jan 26. Int J Parasitol. 2019. PMID: 30690089 Free PMC article. Review.
-
Sexual Development in Non-Human Parasitic Apicomplexa: Just Biology or Targets for Control?Animals (Basel). 2021 Oct 4;11(10):2891. doi: 10.3390/ani11102891. Animals (Basel). 2021. PMID: 34679913 Free PMC article. Review.
-
Identification of proteins expressed by Babesia bigemina kinetes.Parasit Vectors. 2019 May 28;12(1):271. doi: 10.1186/s13071-019-3531-7. Parasit Vectors. 2019. PMID: 31138276 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

