Cloning and Phylogenetic Analysis of Brassica napus L. Caffeic Acid O-Methyltransferase 1 Gene Family and Its Expression Pattern under Drought Stress
- PMID: 27832102
- PMCID: PMC5104432
- DOI: 10.1371/journal.pone.0165975
Cloning and Phylogenetic Analysis of Brassica napus L. Caffeic Acid O-Methyltransferase 1 Gene Family and Its Expression Pattern under Drought Stress
Abstract
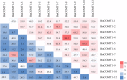
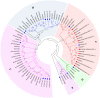
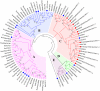
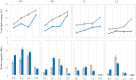
For many plants, regulating lignin content and composition to improve lodging resistance is a crucial issue. Caffeic acid O-methyltransferase (COMT) is a lignin monomer-specific enzyme that controls S subunit synthesis in plant vascular cell walls. Here, we identified 12 BnCOMT1 gene homologues, namely BnCOMT1-1 to BnCOMT1-12. Ten of 12 genes were composed of four highly conserved exons and three weakly conserved introns. The length of intron I, in particular, showed enormous diversification. Intron I of homologous BnCOMT1 genes showed high identity with counterpart genes in Brassica rapa and Brassica oleracea, and intron I from positional close genes in the same chromosome were relatively highly conserved. A phylogenetic analysis suggested that COMT genes experience considerable diversification and conservation in Brassicaceae species, and some COMT1 genes are unique in the Brassica genus. Our expression studies indicated that BnCOMT1 genes were differentially expressed in different tissues, with BnCOMT1-4, BnCOMT1-5, BnCOMT1-8, and BnCOMT1-10 exhibiting stem specificity. These four BnCOMT1 genes were expressed at all developmental periods (the bud, early flowering, late flowering and mature stages) and their expression level peaked in the early flowering stage in the stem. Drought stress augmented and accelerated lignin accumulation in high-lignin plants but delayed it in low-lignin plants. The expression levels of BnCOMT1s were generally reduced in water deficit condition. The desynchrony of the accumulation processes of total lignin and BnCOMT1s transcripts in most growth stages indicated that BnCOMT1s could be responsible for the synthesis of a specific subunit of lignin or that they participate in other pathways such as the melatonin biosynthesis pathway.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

