Secreted Effectors Encoded within and outside of the Francisella Pathogenicity Island Promote Intramacrophage Growth
- PMID: 27832588
- PMCID: PMC5384264
- DOI: 10.1016/j.chom.2016.10.008
Secreted Effectors Encoded within and outside of the Francisella Pathogenicity Island Promote Intramacrophage Growth
Abstract
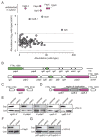
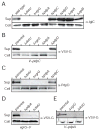
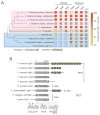
The intracellular bacterial pathogen Francisella tularensis causes tularemia, a zoonosis that can be fatal. The type VI secretion system (T6SS) encoded by the Francisella pathogenicity island (FPI) is critical for the virulence of this organism. Existing studies suggest that the complete repertoire of T6SS effectors delivered to host cells is encoded by the FPI. Using a proteome-wide approach, we discovered that the FPI-encoded T6SS exports at least three effectors encoded outside of the island. These proteins share features with virulence determinants of other pathogens, and we provide evidence that they can contribute to intramacrophage growth. The remaining proteins that we identified are encoded within the FPI. Two of these FPI-encoded proteins constitute effectors, whereas the others form a unique complex required for core function of the T6SS apparatus. The discovery of secreted effectors mediating interactions between Francisella and its host significantly advances our understanding of the pathogenesis of this organism.
Copyright © 2016 Elsevier Inc. All rights reserved.
Figures



Similar articles
-
Francisella tularensis IglG Belongs to a Novel Family of PAAR-Like T6SS Proteins and Harbors a Unique N-terminal Extension Required for Virulence.PLoS Pathog. 2016 Sep 7;12(9):e1005821. doi: 10.1371/journal.ppat.1005821. eCollection 2016 Sep. PLoS Pathog. 2016. PMID: 27602570 Free PMC article.
-
Lipidation of the FPI protein IglE contributes to Francisella tularensis ssp. novicida intramacrophage replication and virulence.Pathog Dis. 2014 Oct;72(1):10-8. doi: 10.1111/2049-632X.12167. Epub 2014 May 8. Pathog Dis. 2014. PMID: 24616435 Free PMC article.
-
A response regulator promotes Francisella tularensis intramacrophage growth by repressing an anti-virulence factor.Mol Microbiol. 2016 Aug;101(4):688-700. doi: 10.1111/mmi.13418. Epub 2016 Jun 10. Mol Microbiol. 2016. PMID: 27169554 Free PMC article.
-
Molecular and genetic basis of pathogenesis in Francisella tularensis.Ann N Y Acad Sci. 2007 Jun;1105:138-59. doi: 10.1196/annals.1409.010. Epub 2007 Mar 29. Ann N Y Acad Sci. 2007. PMID: 17395737 Review.
-
The Francisella Type VI Secretion System.Front Cell Infect Microbiol. 2018 Apr 23;8:121. doi: 10.3389/fcimb.2018.00121. eCollection 2018. Front Cell Infect Microbiol. 2018. PMID: 29740542 Free PMC article. Review.
Cited by
-
Breaking the cellular defense: the role of autophagy evasion in Francisella virulence.Front Cell Infect Microbiol. 2024 Dec 24;14:1523597. doi: 10.3389/fcimb.2024.1523597. eCollection 2024. Front Cell Infect Microbiol. 2024. PMID: 39776438 Free PMC article.
-
A novel stabilization mechanism for the type VI secretion system sheath.Proc Natl Acad Sci U S A. 2021 Feb 16;118(7):e2008500118. doi: 10.1073/pnas.2008500118. Proc Natl Acad Sci U S A. 2021. PMID: 33558227 Free PMC article.
-
Outer membrane tube formation by Francisella novicida involves extensive envelope modifications and is linked with type VI secretion and alterations to the host phagosomal membrane.mBio. 2025 Jun 11;16(6):e0106025. doi: 10.1128/mbio.01060-25. Epub 2025 May 19. mBio. 2025. PMID: 40387340 Free PMC article.
-
Phenotypic and transcriptional characterization of F. tularensis LVS during transition into a viable but non-culturable state.Front Microbiol. 2024 Feb 6;15:1347488. doi: 10.3389/fmicb.2024.1347488. eCollection 2024. Front Microbiol. 2024. PMID: 38380104 Free PMC article.
-
Atomic Structure and Phospholipid Binding Properties of the Francisella Type VI Secretion System Effector Protein PdpC.bioRxiv [Preprint]. 2025 May 8:2025.05.08.652963. doi: 10.1101/2025.05.08.652963. bioRxiv. 2025. PMID: 40654996 Free PMC article. Preprint.
References
-
- Aasland R, Abrams C, Ampe C, Ball LJ, Bedford MT, Cesareni G, Gimona M, Hurley JH, Jarchau T, Lehto VP, et al. Normalization of nomenclature for peptide motifs as ligands of modular protein domains. FEBS letters. 2002;513:141–144. - PubMed
-
- Barrett AJ, Rawlings ND. Evolutionary lines of cysteine peptidases. Biol Chem. 2001;382:727–733. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

