A Herpesvirus Protein Selectively Inhibits Cellular mRNA Nuclear Export
- PMID: 27832591
- PMCID: PMC5130111
- DOI: 10.1016/j.chom.2016.10.004
A Herpesvirus Protein Selectively Inhibits Cellular mRNA Nuclear Export
Abstract
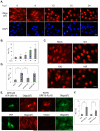
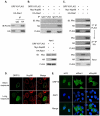
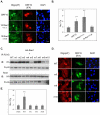
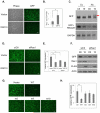
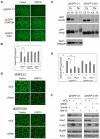
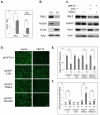
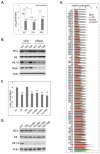
Nuclear mRNA export is highly regulated to ensure accurate cellular gene expression. Viral inhibition of cellular mRNA export can enhance viral access to the cellular translation machinery and prevent anti-viral protein production but is generally thought to be nonselective. We report that ORF10 of Kaposi's sarcoma-associated herpesvirus (KSHV), a nuclear DNA virus, inhibits mRNA export in a transcript-selective manner to control cellular gene expression. Nuclear export inhibition by ORF10 requires an interaction with an RNA export factor, Rae1. Genome-wide analysis reveals a subset of cellular mRNAs whose nuclear export is blocked by ORF10 with the 3' UTRs of ORF10-targeted transcripts conferring sensitivity to export inhibition. The ORF10-Rae1 interaction is important for the virus to express viral genes and produce infectious virions. These results suggest that a nuclear DNA virus can selectively interfere with RNA export to restrict host gene expression for optimal replication.
Keywords: 3′ UTR; KSHV; Nup98; ORF10; Rae1; herpesvirus; late gene; mRNA nuclear export.
Published by Elsevier Inc.
Figures
Similar articles
-
Molecular mechanism underlying selective inhibition of mRNA nuclear export by herpesvirus protein ORF10.Proc Natl Acad Sci U S A. 2020 Oct 27;117(43):26719-26727. doi: 10.1073/pnas.2007774117. Epub 2020 Oct 8. Proc Natl Acad Sci U S A. 2020. PMID: 33033226 Free PMC article.
-
SARS-CoV-2 ORF6 Disrupts Bidirectional Nucleocytoplasmic Transport through Interactions with Rae1 and Nup98.mBio. 2021 Apr 13;12(2):e00065-21. doi: 10.1128/mBio.00065-21. mBio. 2021. PMID: 33849972 Free PMC article.
-
Recruitment of the complete hTREX complex is required for Kaposi's sarcoma-associated herpesvirus intronless mRNA nuclear export and virus replication.PLoS Pathog. 2008 Oct;4(10):e1000194. doi: 10.1371/journal.ppat.1000194. Epub 2008 Oct 31. PLoS Pathog. 2008. PMID: 18974867 Free PMC article.
-
Kaposi's sarcoma-associated herpesvirus ORF57 protein: exploiting all stages of viral mRNA processing.Viruses. 2013 Jul 26;5(8):1901-23. doi: 10.3390/v5081901. Viruses. 2013. PMID: 23896747 Free PMC article. Review.
-
Inhibition of Host Gene Expression by KSHV: Sabotaging mRNA Stability and Nuclear Export.Front Cell Infect Microbiol. 2021 Apr 9;11:648055. doi: 10.3389/fcimb.2021.648055. eCollection 2021. Front Cell Infect Microbiol. 2021. PMID: 33898329 Free PMC article. Review.
Cited by
-
When Poly(A) Binding Proteins Meet Viral Infections, Including SARS-CoV-2.J Virol. 2022 Apr 13;96(7):e0013622. doi: 10.1128/jvi.00136-22. Epub 2022 Mar 16. J Virol. 2022. PMID: 35293770 Free PMC article. Review.
-
Transcriptional and post-transcriptional regulation of viral gene expression in the gamma-herpesvirus Kaposi's sarcoma-associated herpesvirus.Curr Clin Microbiol Rep. 2018 Dec;5(4):219-228. doi: 10.1007/s40588-018-0102-1. Epub 2018 Aug 3. Curr Clin Microbiol Rep. 2018. PMID: 30854283 Free PMC article.
-
Persistent Replication of a Chikungunya Virus Replicon in Human Cells Is Associated with Presence of Stable Cytoplasmic Granules Containing Nonstructural Protein 3.J Virol. 2018 Jul 31;92(16):e00477-18. doi: 10.1128/JVI.00477-18. Print 2018 Aug 15. J Virol. 2018. PMID: 29875241 Free PMC article.
-
Atypical nuclear envelope condensates linked to neurological disorders reveal nucleoporin-directed chaperone activities.Nat Cell Biol. 2022 Nov;24(11):1630-1641. doi: 10.1038/s41556-022-01001-y. Epub 2022 Oct 27. Nat Cell Biol. 2022. PMID: 36302970 Free PMC article.
-
Viral dUTPases: Modulators of Innate Immunity.Biomolecules. 2022 Jan 28;12(2):227. doi: 10.3390/biom12020227. Biomolecules. 2022. PMID: 35204728 Free PMC article. Review.
References
-
- Blevins MB, Smith AM, Phillips EM, Powers MA. Complex formation among the RNA export proteins Nup98, Rae1/Gle2, and TAP. Journal of Biological Chemistry. 2003;278:20979–20988. - PubMed
-
- Brown JA, Bharathi A, Ghosh A, Whalen W, Fitzgerald E, Dhar R. A mutation in the Schizosaccharomyces pombe rae1 gene causes defects in poly(A)+ RNA export and in the cytoskeleton. The Journal of biological chemistry. 1995;270:7411–7419. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

